
International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056
Volume: 12 Issue: 06 | Jun 2025 www.irjet.net p-ISSN:2395-0072


International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056
Volume: 12 Issue: 06 | Jun 2025 www.irjet.net p-ISSN:2395-0072
Mr. N. L. Bhale¹, Prasad Navale², Om Jadhav³, Sakshi Dobhal´, Rohit Jadhavµ
1 Head of Department, Department of Information Technology, Matoshri College of Engineering & Research Centre, Eklahare, Maharashtra, India
2,3,4,5 Students, Department of Information Technology, Matoshri College of Engineering & Research Centre, Eklahare, Maharashtra, India
Abstract -Skindiseasesareprevalentworldwide,affecting millions of people and often presenting diagnostic challenges due to their diverse manifestations. Deep learningtechniques haveshownpromise inautomatingthe diagnosis of skin diseases, leveraging large datasets and powerful computational methods. However, the performance of existing models may vary depending on factors such as dataset quality, model architecture, and featureextractionmethods
Key Words: Convolution neural network, combined decision, deep learning, skin cancer.
Skin diseases are a significant public health concern worldwide, affecting individuals of all ages and demographics. The diagnosis and management of these conditionsoftenposechallengesduetothewidevarietyof dermatol(CNNs) have been particularly effective in analyzing staogical manifestations and the need for accurate and timely assessment. While traditional diagnostic methods rely heavily on clinical expertise and histopathological examination, the advent of artificial intelligence (AI) and deep learning techniques has revolutionized the field of dermatology by enabling automatedandefficientskindiseasediagnosis.
Deep learning, a subset of AI, has shown remarkable success in various medical imaging tasks, including dermatology. Convolutional neural networks tic images of skinlesions,achievingperformancecomparabletooreven surpassing that of dermatologists in certain scenarios. However,conventionalstaticimage-basedapproachesmay overlook crucial temporal information inherent in the evolutionofskindiseasesovertime.
Dynamic testing in deep learning offers a promising solution to this limitation by leveraging sequential data to capturetemporaldynamicsinskinlesions.Unlikestatic image analysis, dynamic testing involves the analysis of sequential images taken at different time points, enabling the model to learn from the progression or regression of skin diseases. This temporal perspective can provide valuable insights for disease prediction, monitoring, and treatmentresponseassessment.
Skindiseasesposeasignificantburdenonpublichealth globally, affecting millions of individuals and often requiring timely and accurate diagnosis for effective management. Despite advances in medical imaging and diagnostic techniques, the complexity and variability of dermatologicalconditionspresentchallengesforclinicians in accurately identifying and treating these diseases. Traditional diagnostic methods rely heavily on visual inspection and subjective assessment, which can lead to variabilityindiagnosesanddelaysintreatmentinitiation.
TemporalDynamicsofSkinLesions:Skindiseasesoften exhibit dynamic changes over time, including progression, regression, and response to treatment. By analyzing sequentialimagescapturedatdifferenttimepoints,wecan gain valuable insights into the temporal evolution of skin lesions, which may provide important diagnostic and prognosticinformation.
Improved Diagnostic Accuracy: Conventional static image- based approaches may overlook critical temporal patterns and dynamics present in skin lesions. Dynamic testing in deep learning allows us to leverage sequential data to capture these temporal dynamics, potentially leading to more accurate and reliable disease predictions comparedtostaticimageanalysisalone.
The purpose of the proposed system, "Skin Disease Prediction using Dynamic Testing in Deep Learning," is to developanadvancedcomputationaltoolthatleveragesthe temporaldynamicsofskinlesionstoenhancetheaccuracy, efficiency, and automation of skin disease diagnosis and prediction. The system aims to address the following key objectives:
1. Improved Diagnostic Accuracy: By analyzing sequential images of skin lesions captured at different time points, the system seeks to capture temporal patterns and dynamics that may contain valuable diagnostic information. Byincorporatingtemporalinformationintothe predictionprocess, thesystemaimstoimprove the accuracy of skin disease diagnosis

International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056
Volume: 12 Issue: 06 | Jun 2025 www.irjet.net p-ISSN:2395-0072
compared to traditional static image-based approaches.
2. Real-Time Disease Monitoring: The system enables real-time monitoring of disease progression, regression, and treatment response by analyzing sequential images over time. By tracking changes in skin lesions, clinicians can obtain timely insights into disease dynamics and treatment effectiveness, facilitating proactive management and intervention
Current diagnostic methods for skin diseases may lack accuracy,especiallyincaseswherethediseasemanifestsas subtle changes over time. Static image-based approaches often fail to capture the dynamic evolution of skin lesions, leading to diagnostic errors and misclassification of diseases.
Diagnostic delays in skin diseases can have significant implications for patient outcomes, leading to prolonged suffering, disease progression, and potentially irreversible damage. Traditional diagnostic workflows may involve time- consuming manual assessment, resulting in delayed diagnosisandtreatmentinitiation.
Skin disease prediction using dynamic testing within deep learning frameworks represents a cutting-edge approach poised to revolutionize dermatological diagnostics. By harnessing the temporal evolution of skin lesions captured through sequential imaging, this methodology aims to provide more accurate and timely predictions of disease progression and regression. Recent researchinthisdomainhasseensignificantadvancements, withstudiesexploringtheeffectivenessofrecurrentneural networks (RNNs) and temporal convolutional networks (TCNs) in analyzing sequential skin images. These models treat the temporal sequence of skinimages as informative datastreams,allowingthemtocapturedynamicchangesin lesion morphology over time. Additionally, attention mechanisms have been integrated into deep learning architectures to dynamically allocate focus to relevant regions within sequential images, further enhancing predictive accuracy. Such methodologies have demonstrated promising results in accurately predicting disease dynamics and tracking lesion evolution, thus offering invaluable insights for clinical decision-making and patient management. Overall, skin disease prediction using dynamic testing in deep learning holds immense potential for improving diagnostic accuracy, treatment planning,and patientoutcomesindermatology.
The proposed system aims to develop an advanced computational tool that leverages dynamic testing within deep learning frameworks for accurate and efficient prediction of skin diseases. By analyzing the temporal evolution of skin lesions captured through sequential imaging, the system seeks to enhance diagnostic accuracy, automate diagnostic workflows, and improve treatment planningindermatology.
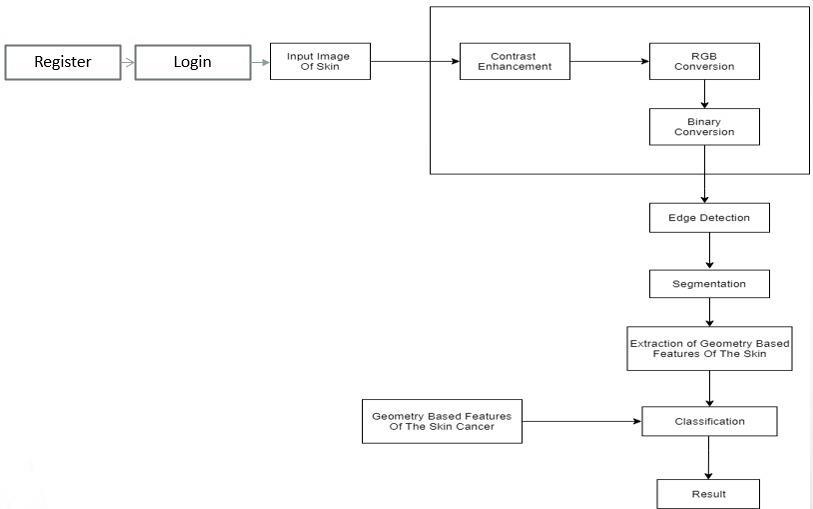
Figure1:SystemArchitecture
This module is responsible for collecting sequential skin images along with temporal annotations from various sources such as medical databases or imaging devices. Preprocessing techniques are applied to standardize image resolution, remove noise, and augment the dataset to enhancemodelrobustness.Temporalinformationsuch as timestamps, lesion characteristics, and patient metadataareincorporatedintothedataset
A. Image Database: The database is downloaded from publicly available images on the ISIC website. The database included the ISBI-2016 challenge which has RGB dermoscopic images along with their labels andsegmentationgroundtruths.
B. Pre-Processing As dataset images have different artifacts, so pre-processing is done on them to make them more meaningful. Pre-processing included image resizing, noise removal, contrast stretching, RGB to Gray conversion, and hair removal. For contrast enhancement, a new method is proposed. Contrast enhancement significantly improves the

International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056
Volume: 12 Issue: 06 | Jun 2025 www.irjet.net p-ISSN:2395-0072
results of segmentation in the next phase. Proposed contrast stretching is based on the mean and standard deviation of pixels intensities of images. Minimumandmaximumintensityvaluesi.e."Lowin" and "High in" of input images values of input images arecalculated
1.IncreaseDatasetSize
2.EfficiencyofAlgorithms
3.AddMoreDiseases
4.FeatureEngineering
5.ModelEvaluationMetrics
6.Interpretability
7.ClinicalValidation
6. PROBLEM SOLVING
1.LimitedDataset:Therelianceonasingledataset,ISICISBI2016,maylimitthegeneralizabilityoftheproposed method. It's important to validate the approach on multiple datasets to ensure its robustness across differentpopulationsandimagingconditions.
2.Class Imbalance: Although the paper addresses class imbalance using the SMOTE technique, this may introduce synthetic data that could affect the generalization of the model. Careful consideration and evaluationoftheimpactofclassbalancingtechniqueson modelperformancearenecessary.
3.Feature Selection: While the paper proposes a wrapper- based approach for feature selection, the choiceof95features out of the initial 149 featuresmay not be optimal. Further exploration of feature selection techniques and their impact on classification performancecouldbebeneficial.
4.Evaluation Metrics: The paper primarily focuses on classification accuracy as the evaluation metric. However, other metrics such as sensitivity, specificity, precision, and F1 score provide a more comprehensive understanding of model performance, especially in medical applications where false positives and false negativeshavedifferentconsequences.
5.Model Interpretability: While Random Forest classifiersofferhighaccuracy,theyareoftenconsidered as black-box models, making it challenging to interpret the underlying decision-making process. Providing insights into the features contributing to classification decisions could enhance the interpretability of the model.
6.Clinical Validation: The proposed method needs to undergo clinical validation to assess its real-world effectiveness in assisting dermatologists in skin cancer diagnosis. Clinical trials or studies involving dermatologists'feedbackandvalidationonindependent datasets would provide valuable insights into the practicalutilityofthesystem.
7. Computational Resources: The computational requirements for training and deploying machine learning models, especially Random Forest, may be significant. Consideration of computational efficiency and scalability is essential for real-world deployment, especiallyinresource-constrainedsettings
Addressing these issues would further enhance the robustness, reliability, and practical applicability of the proposedskincancerclassificationsystem.
7. PROPOSE SYSTEM
7.1Dashboard
7.2 SkinDiseaseDetection
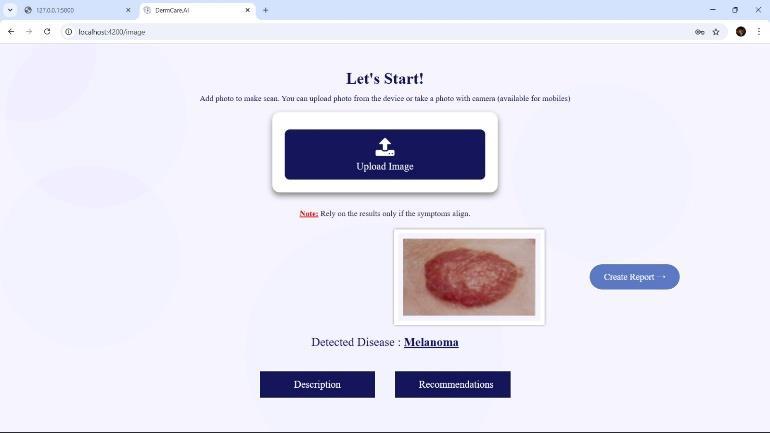
Figure3:SkinDiseaseDetection

International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056
Volume: 12 Issue: 06 | Jun 2025 www.irjet.net p-ISSN:2395-0072
8. PROPOSE SYSTEM FLOWCHART
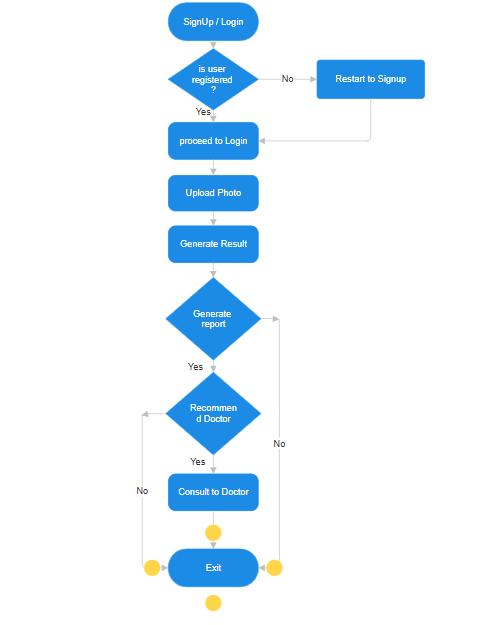
Figure5:ProposeSystemFlowchart
9.CONCLUSION
In this project, different phases of image processing were applied on skin Nodules. From these different image processing techniques, the fuzzy filter will provide the efficient de noising. Segmentation done by marker-based watershed algorithm, givesvarious region of image.GLCM is used to ex- tract the different features of image, and which takes less time for generating the result. These results are passed through CNN Classifier, which classifies the nodules as benign or malignant. The proposed model
performs better than individual learners with respect to differentqualitymeasuresi.e.sensitivity,accuracy,F-Score, specificity, false-Positive, and precision. In future, we are intended to study the achievement of reinforcement learning-basedtechniquesforskincancerdetection.
The project scope for skin cancer prediction involves the development of an advanced and accurate system for the early detection and prediction of skin cancer, encompassing both malignant melanoma and nonmelanoma types. The primary objective is to create a comprehensivesolutionthatenhancesdiagnosticaccuracy, ensures personalized predictions based on individual patient characteristics, minimizes reliance on invasive diagnosticprocedures,andenablescontinuousmonitoring of skin health for proactive management. The project will leverage diverse datasets, including images, dermoscopic images, and patient history data, to train deep learning models that incorporate multiple diagnostic techniques. The system will feature an intuitive user interface to facilitateseamlessinteractionforhealthcareprofessionals, ensuring accessibility and ease of use. Performance metrics, such as diagnostic accuracy and response time, will be established to measure the effectiveness of the system. Ethical considerations, security measures, and compliance with healthcare regulations will be integral componentsoftheproject.Thedeliverablesincludeafully functional skin cancer prediction system, comprehensive documentation, and considerations for future scalability and research opportunities. Through this project, we aim to significantly contribute to early detection, personalized medicine, and improved healthcare outcomes in the domainofskincancer.
[1] M.R.Foundation.(2016). Melanoma is the Deadliest Form of Skin Cancer. [Online]. Available: https://melanoma.org/
[2] Z. Waheed, A. Waheed, M. Zafar, and F. Riaz, ‘‘An efficient machine learning approach for the detection of melanomausingdermoscopicimages,’’inProc.Int.Conf. Commun., Comput. Digit. Syst. (C-CODE), Mar. 2017, pp. 316–319.
[3] M. Sattar and A. Majid, ‘‘Lung cancer classification modelsusingdiscriminantinformationofmutatedgenes in protein amino acids sequences,’’ Arabian J. Sci. Eng., vol.44,no.4,pp.3197–3211,Apr.2019.
[4] A.Majid,S.Ali,M.Iqbal,andN.Kausar,‘‘Predictionof human breast and colon cancers from imbalanced data using nearest neighbor and support vector machines,’’ Comput. Methods Programs Biomed., vol. 113, no. 3, pp. 792–808,Mar.2014.

International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056
Volume: 12 Issue: 06 | Jun 2025 www.irjet.net p-ISSN:2395-0072
[5] R. Seeja and A. Suresh, ‘‘Deep learning based skin lesion segmentation and classification of melanoma using support vector machine (SVM),’’ Asian Pacific J. CancerPrevention,vol.20,no.5,p.1555,Feb.2019.
[6] L.Bi,J.Kim,E.Ahn,A.Kumar,M.Fulham,andD.Feng, ‘‘Dermoscopic image segmentation via multistage fully convolutional networks,’’ IEEE Trans. Biomed. Eng., vol. 64,no.9,pp.2065–2074,Sep.2017.
[7] U.-O. Dorj, K.-K. Lee, J.-Y. Choi, and M. Lee, ‘‘The skin cancer classification using deep convolutional neural network,’’ Multimedia Tools Appl., vol. 77, no. 8, pp. 9909–9924,Apr.2018.
[8] A. Esteva, B. Kuprel, R. A. Novoa, J. Ko, S. M. Swetter, H. M. Blau, and S. Thrun ‘‘Dermatologist-level classificationofskincancerwithdeepneuralnetworks,’’ Nature,vol.542,no.7639,pp.115–118,2017.
[9] Kieffer, B., Babaie, M., Kalra, S & Tizhoosh, H. Convolutional neural networks for histopathology image classification:Training vs. using pre-trained networks. arXivpreprintarXiv:1710.05726(2017)
[10] G. Haung, Z.Liu, K. Q. Weinberger, and L. Maaten. Densely connected convolutional networks. In CVPR, 2017\
[11] P. Tschandl, C. Rosendhal, and H. Kittler, The HAM10000 dataset, a large collection of multi-soruce dermatoscopic images of common pigmented skin lesions, Sci. Data, vol. 5, p. 180161, 2018 (download:https://dataverse.harvard.edu/dataset.xhtml? persistentId=doi:10.7910/DVN/DBW86T)