
International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056
Volume: 12 Issue: 05 | May 2025 www.irjet.net p-ISSN: 2395-0072


International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056
Volume: 12 Issue: 05 | May 2025 www.irjet.net p-ISSN: 2395-0072
Mr. Shubham Paliwal, Mr. Abhishek Purohit
Student of B.Tech Computer Science and Engineering, Bikaner Technical University, Bikaner, Rajasthan, India
Assistant Professor, Department of Computer Science, Bikaner Technical University, Bikaner, Rajasthan, India
Abstract - Agriculture forms the backbone of many economies,yetfarmersoftenstruggletodetectplantdiseases early,leadingtosignificantcroplosses.Recentadvancements inartificialintelligencehaveshownpromisingapplicationsin agriculture, especially in disease diagnosis using image processing. This paper introduces PlantoScope, a convolutionalneuralnetwork(CNN)-basedmobileapplication designed for real-time plant disease diagnosis through leaf images. The model is trained on the PlantVillage dataset comprising 38 distinct plant classes and diseases. The application allows users to capture or upload leaf images, which are then analyzed by a deep learning model deployed via a Flask API. The output includes the plant type, disease name,cause,andrecommendedcure.Thesystemachieves an accuracy of 96% using a .h5 model and 94% using a compressed .tflite version, making it viable for mobile deployment. This solution is designed to be scalable and farmer-friendly,withplanstointegratethermalimagingand a binary leaf classifier in the future.
KeyWords: Plantdiseasedetection,ConvolutionalNeural Network (CNN), Mobile application, Deep learning, Flask API, TensorFlow, Agriculture, PlantVillage, Image classification
Agriculture is a vital sector in many developing nations, where crop health directly affects economic stability and food security. Early detection of plant diseases plays a critical role in ensuring high crop yields and minimizing losses. Traditionally, farmers rely on visual inspection or expertadvice,whichmaybesubjective,time-consuming,and inaccessibleinremoteregions.
With the rise of deep learning and image processing, automatedplantdiseasedetectionhasbecomeapromising alternative. Convolutional Neural Networks (CNNs) have provenparticularlyeffectiveforimageclassificationtasks, includingplantpathology.
This paper presents PlantoScope, a mobile-based plant diseasediagnosisapplicationpoweredbyaCNNmodel.By capturingoruploadinganimageofaleaf,usersreceivean instantpredictionoftheplanttype,diseasename,possible cause,andrecommendedcure.Thebackendmodelistrained onthewell-knownPlantVillagedataset,andtheapplication isoptimizedfordeploymentusingTensorFlowLite.
PlantoScopeaimstobridgethegapbetweenmodernAItools and the needs of farmers by offering an easy-to-use, realtimediseaseidentificationsystem.
Recentyearshavewitnessedsignificantadvancementsinthe use of deep learning for agricultural applications. Several studies have demonstrated the efficacy of Convolutional NeuralNetworks(CNNs)inplantdiseaseidentificationusing image-baseddatasets.
Mohanty et al. (2016) were among the first to use deep learningmodelsonthePlantVillagedataset,achievingover 99% accuracy on test data using AlexNet and GoogLeNet architectures. Their work validated the potential of image classificationmodelsforfield-leveldiseasedetection.
Ferentinos(2018)furtherextendedthislineofresearchby evaluating multiple CNN architectures, including VGG and AlexNet,acrossseveralplantspeciesanddiseasecategories. His study confirmed CNNs’ robustness even in complex classificationtasksinvolvingmultipleclassesandsubtleleaf texturevariations.
However,mostoftheexistingresearchhasfocusedonmodel accuracy in offline environments. Limited work has been done to translate these models into lightweight, real-time mobile applications accessible to end-users, particularly farmers.
This gap in deployment-focused research forms the motivation for PlantoScope, which not only achieves high classification accuracy but also delivers the functionality through a user-friendly mobile interface with TensorFlow Liteintegration.
The proposed model was trained on the publicly available PlantVillage datasetsourcedfromKaggle.Itconsistsof 54,305 images of 14 different plants across 38 classes, each representingaspecificcombinationofplanttypeanddisease condition, such as Apple___Black_rot, Tomato___Leaf_Mold, andCorn_(maize)___Common_rust_.
Itconsistsofimagesof14differentplants,namely-Tomato, Strawberry.Squash,Soybean,Raspberry,Potato,BellPepper, Peach,Orange,Grape,Corn(Maize),Cherry,Blueberry,Apple

International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056
Volume: 12 Issue: 05 | May 2025 www.irjet.net p-ISSN: 2395-0072
To ensure compatibility and efficiency in training, the followingpreprocessingstepswereapplied:
Resizing:Allimageswereresizedto 224×224pixels tomatchtheinputdimensionsoftheCNNmodel.
Normalization: Pixel values were scaled to the range [0, 1] bydividingby255.
Label Encoding:Classfoldernameswereconverted to one-hot encoded labels using ImageDataGeneratorwithcategoricalmode.
Dataset Split: The data was divided into 80% training and 20%validation.Aseparatetestdataset wasusedforfinalevaluation.
Shuffling: Disabled during test time to preserve labelorderforconfusionmatrixanalysis.
The proposed model follows a sequential Convolutional Neural Network (CNN) architecture optimized for image classificationtasks.Thedesignwaskeptintentionallysimple toreducecomplexitywhileachievinghighaccuracyonthe PlantVillagedataset.
4.1 Architecture Details
TheCNNconsistsofthefollowinglayers:
Conv2D Layer (32 filters, 3×3 kernel, ReLU): Captureslow-levelfeaturessuchasedgesandtextures.
MaxPooling2D (2×2): Reducesspatialdimensionsand computationalload.
Conv2D Layer (64 filters, 3×3 kernel, ReLU): Extracts higher-levelfeaturesfrompooledoutputs.
MaxPooling2D(2×2):Furtherreducesfeaturemapsize.
Flatten Layer: Transformsthe3Dfeaturemapsintoa 1Dvector.
Dense Layer (256 units, ReLU): Learns complex patternsfromflattenedfeatures.
Output Dense Layer (38units,Softmax):Outputsclass probabilitiesfor38categories.
4.2 Model Summary
Themodelhas 47.8M parametersintotal.Thesummaryof themodel,describingtheOutputShapeandtheNumberof ParamtersofeachLayerTypeareasfollows:
Layer
Table -1: ModelSummary
Conv2D (222,222,32) 896
MaxPooling2D (111,111,32) 0
Conv2D (109,109,64) 18,496
MaxPooling2D (54,54,64) 0
Flatten (186,624) 0
Dense (256) 47,776,000
Dense (38) 9,766
5.1 Data Flow Diagrams
The Data Flow Diagrams (DFDs) of PlantoScope provide a visualrepresentationofhowdatamovesthroughthesystem.

Fig -1: Level0DFD
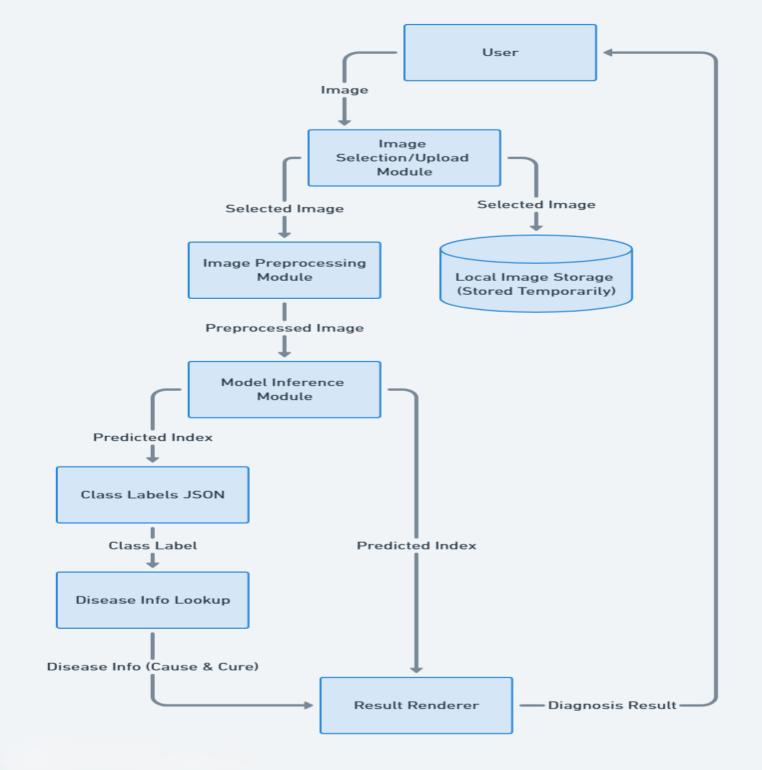
Fig -2:Level1DFD

International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056
Volume: 12 Issue: 05 | May 2025 www.irjet.net p-ISSN: 2395-0072
Thefollowingsystemflowdiagramillustratesthesequence ofoperationsandlogicwithinthe PlantoScope application.It highlights key user interactions such as image capture or selection from gallery, preprocessing (including resizing, normalizingpixelvalues)toprepareformodelprediction and image submission to the backend API, followed by authentication of the request by comparing the API key received in the request body with the API key set on the backend server, model inference using the CNN-based classifier, retrieval of prediction results including disease name,cause,andcure,andfinally,displayingthediagnosis alongwiththeselectedimageontheresultscreen.
TheSystemFlowDiagramisasfollows:
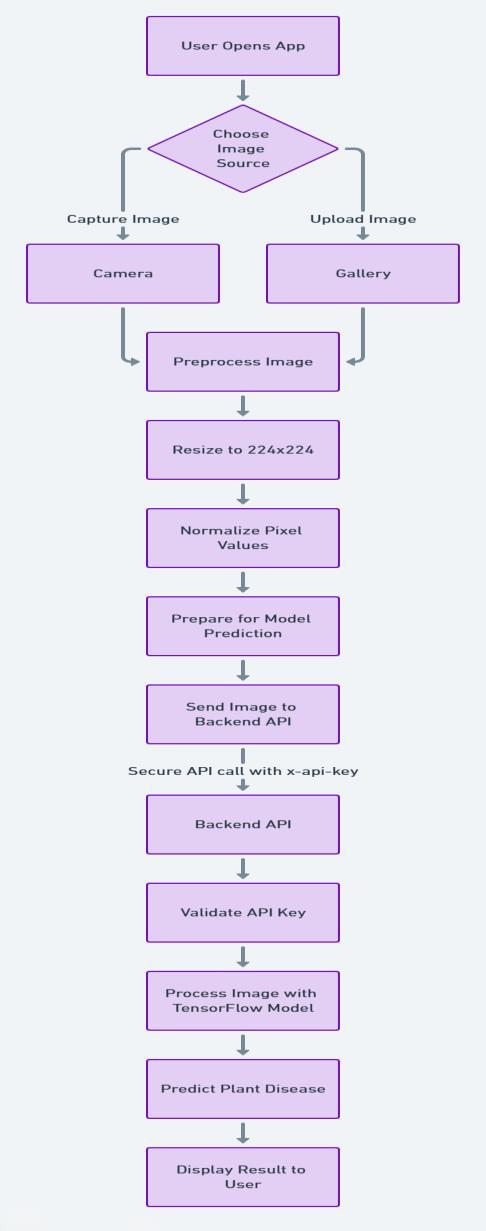
6.1
ThePlantVillagedatasetwasdividedinto:
Training Set: 80%ofimages
Validation Set: 10%ofimages
Test Set: 10%ofimages
Stratified splitting was used to maintain class distribution acrosssplits
6.2 Training Parameters
Optimizer: Adam
Loss Function: CategoricalCrossentropy
Batch Size: 32
Epochs: 20
Learning Rate: 0.001(withdecay)
Early Stopping: Enabled(patience=3)
6.3 Evaluation Metrics
To comprehensively evaluate model performance, the followingmetricswereused:
Accuracy
Precision
Recall
F1-Score
Confusion Matrix
Thesemetrics were computedonthetestsetusing scikitlearn.
To enable mobile deployment, the trained .h5 model was converted to TensorFlow Lite (.tflite) format without quantization,preservingperformanceforinferenceonevery type of mobile devices, even on those having low-end specifications, because the server’s computation power is beingutilizedtorunthemodelinferenceviaanAPIcall,the mobileisonlyusedtoprocesstheUItasks.

International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056
Volume: 12 Issue: 05 | May 2025 www.irjet.net p-ISSN: 2395-0072
7.1 Model Performance
The trained CNN model achieved high accuracy and reliability across the 38 plant disease classes. Below is a summaryoftheevaluationonthetestdataset:
Accuracy: 96%
Precision (macro avg): 96%
Recall (macro avg): 94%
F1-Score (macro avg): 95%
Number of Test Images: 54,305
Number of Classes: 38
7.2 Confusion Matrix Analysis
The confusion matrix confirmed strong classification capability,especiallyforcommondiseasessuchasTomato YellowLeafCurlVirusandCornRust.Someconfusionwas noted between visually similar classes like "Tomato Early Blight" and"TomatoLateBlight",which may benefit from additionalfeatureseparationortrainingdata.
The model was successfully deployed via a Flask API and integrated into a React Nativeapp.Inference resultswere returnedinreal-timewithsatisfactoryperformanceevenon low-endmobilephones.
8.1 Conclusion
Thispaperpresents PlantoScope,alightweightCNN-based mobile application capable of detecting 38 plant diseases usingleafimageclassification.Themodelwastrainedonthe PlantVillagedatasetandachievedahighaccuracyof 96%.It wassuccessfullyconvertedintoaTensorFlowLiteformat, deployedusingaFlask-basedAPI,andintegratedintoauserfriendlymobileinterfacebuiltwithReactNative.Thesystem demonstrates effective real-world usability for farmers, students,andagriculturalresearchersbyprovidingdisease type,cause,andcuresuggestionswithminimallatency.
8.2
While the current results are promising, several enhancementsareplanned:

7.3 .h5 vs .tflite Comparison
Although the .tflite model was converted without quantizationtoretainperformance,aminordropinaverage precision (~2%) was observed, likely due to internal representationdifferencesinTensorFlowLite.However,this tradeoffisacceptablegiventhesizeandspeedadvantages formobiledeployment.
Leaf vs Non-leaf Classifier: To reduce false positives, a preliminary binary classification step canbeaddedtoensuretheuploadedimagecontains aleaf.
Thermal Image Integration: A secondary model trained on thermal imagery may improve disease detection in early stages, especially for temperature-sensitivecrops.
Extended Dataset Support: Incorporatingimages ofIndianplantvarietiesandexpandingthedisease coveragewillfurtherimprovegeneralization.
Multi-language Support: AddingsupportforHindi and regional languages can make the app more accessibletolocalfarmers.
Play / App Store Deployment: Theappisalready testedandreadyforAPKgeneration;publishingit totheGooglePlayStore/AppleAppStoreispartof thefutureplan.
Web / Desktop Deployment: Sincethefrontendis builtusingReactNativewithExpo,itofferscrossplatform flexibility. A web or desktop version of PlantoScopecanbedeployedusingframeworkslike ReactNativeWeb,Electron,oranyotherfrontend byintegratingitwithPlantoScopeAPI,toenhance accessibilityforuserswithoutsmartphones.

International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056
Volume: 12 Issue: 05 | May 2025 www.irjet.net p-ISSN: 2395-0072
Itakethisopportunitytoexpressmygratitudetoallthose peoplewhohavebeendirectlyandindirectlyinvolvedwith me during the completion of this project. I extend my heartfelt thanks to Mr. Abhishek Purohit, Assistant Professor, Department of Computer Science, Bikaner Technical University, Bikaner, Rajasthan, India whose guidance and expertise in ideation, research, and developmenthavebeeninvaluable.Hissupporthashelped menavigatethroughcriticaltimesduringthisproject.Ialso acknowledge with gratitude those who contributed significantly to various aspects of this project. I take full responsibilityforanyremainingoversightsoromissions.
[1] Mohanty,S.P.,Hughes,D.P.,&Salathé,M.(2016). Using deep learning for image-based plant disease detection Frontiers in Plant Science, 7, 1419. https://doi.org/10.3389/fpls.2016.01419
[2] Ferentinos,K.P.(2018). Deep learning models for plant disease detection and diagnosis. Computers and Electronics in Agriculture, 145, 311-318. https://doi.org/10.1016/j.compag.2018.01.009
[3] Hughes, D. P., & Salathé, M. (2015). An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv preprint arXiv:1511.08060. https://arxiv.org/abs/1511.08060
[4] PlantVillage Dataset. https://www.kaggle.com/datasets/abdallahalidev/plan tvillage-dataset
[5] TensorFlow (An end-to-end open source machine learning platform) https://www.tensorflow.org
[6] KerasAPIDocumentation. https://keras.io
[7] Expo & React Native Documentation. https://docs.expo.dev|https://reactnative.dev
[8] FlaskDocumentation. https://flask.palletsprojects.com
[9] Gdown Library (for Google Drive integration). https://github.com/wkentaro/gdown
[10]Python Imaging Library (Pillow). https://pillow.readthedocs.io
[11]Scikit-learn:MachineLearninginPython. https://scikit-learn.org

“Mr. Shubham Paliwal isafinal-year B.Tech student in Computer Science andEngineeringatBikanerTechnical University,Bikaner,Rajasthan,India. Heisapassionatefull-stackdeveloper with proficiency in Java, JavaScript, ReactJS,Firebase,PHP,MySQL,C,C++, Linux, and strong problem-solving skillsgroundedinDataStructuresand Algorithms.
With over three years of hands-on programming experience, he has developed several feature-rich applications,including Chitthi –arealtime chat application, and TrafficTracer – a botnet detection system powered by an XGBoost machine learning model. He actively contributes to open-source communities and consistently exploresinnovativeideasinsoftware development.
More details about his work can be found on his personal website: https://shubhampaliwal.me
He can be reached at shubhampaliwal.dev@gmail.com”

“Mr. Abhishek Purohit is currently serving as an Assistant Professor in theDepartmentofComputerScience andEngineeringatBikanerTechnical University,Bikaner,Rajasthan,India. He holds both M.Tech and B.Tech degrees in Computer Science and Engineeringandispresentlypursuing hisPh.D.
With a strong foundation in programminglanguagessuchasJava, C++, and C, he brings expertise in ideation, research,anddevelopment. Hisprimaryresearchinterestsinclude the application of Artificial IntelligenceandMachineLearningin agriculture,innovationthroughdesign thinking, and developing costeffective,high-impactsolutions.Heis passionate about optimizing current

International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056
Volume: 12 Issue: 05 | May 2025 www.irjet.net p-ISSN: 2395-0072
markettechnologiestocreatescalable innovations that contribute to sustainable development and bring meaningfulchangetosociety. He can be reached at erabhishek.purohit@gmail.com”
© 2025, IRJET | Impact Factor value: 8.315 | ISO 9001:2008 Certified Journal | Page1649