
International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056
Volume: 12 Issue: 05 | May 2025 www.irjet.net p-ISSN:2395-0072


International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056
Volume: 12 Issue: 05 | May 2025 www.irjet.net p-ISSN:2395-0072
N.S. Chidhvii
Reddy1, P.
Deepika Reddy
2, S.
Pranavi
3, Dr. D. Subhashini4
123Student, Electronics and Communication Engineering, MGIT College, Telangana, India
4 Assistant Professor, Electronics and Communication Engineering, MGIT College, Telangana, India
Abstract - India is known for its incredible plant diversity, especially when it comes to medicinal species. For generations, plants have been at the heart of traditional healing practices like Ayurveda, Siddha, and Unani, offering a wide range of natural remedies. Today, as more people turn to nature-based solutions, medicinal plants are becoming increasingly popular as affordable, eco-friendly, and generally safer alternatives to chemical-based medicines. Still, one of the biggest challenges lies in accurately identifying and classifying these plants. With so many species that often look alike, it’s easy to make mistakes even for experts. Misidentifying a plant can lead to using the wrong one, which might not only reduce the treatment’s effectiveness but could also be harmful. This challenge highlights the need for a dependable and easy-touse identification method. In response, this research aims to develop intelligent software that can accurately recognize medicinal plants using image processing and advanced deep learning technologies. By combining computer vision with modern neural networks, the project hopes to make plant identification both faster and more reliable. Such a tool could be incredibly helpful not just for researchers and botanists, but also for everyday users, herbal medicine enthusiasts, and community health workers who rely on the safeandeffective useofmedicinal plants
Keywords- Deeplearning, CNN, Resnet50, VGG16, Softmax, Convolution, ConfidenceScore,
1. INTRODUCTION
India stands out as one of the world’s most biologically richnations,thankstoitsvastandvariedplantlife.Among thisnaturalabundance,medicinalplantshaveheldaplace of importance for centuries, serving as the backbone of traditional healing systems like Ayurveda, Siddha, and Unani. Known for their natural healing properties, these plants are now gaining renewed attention as environmentally friendly and sustainable alternatives to modernpharmaceuticals.
However, to truly benefit from these valuable resources, it’s crucial to accurately identify each plant and understand its specific qualities. Despite the wealth of traditional and modern knowledge available, the large number of plant species many of which look strikingly similar makesmanualidentificationaslow,difficult,and often unreliable task. Mistakes in recognizing the right
plantcanleadtoineffective remediesor evenposehealth risks.
To address these challenges, an innovative software solution is being developed that uses image recognition and advanced deep learning techniques. This intelligent toolaimstotransformhowmedicinalplantsareidentified and used. By analyzing plant images, the system will be able to accurately distinguish between species and provide detailed insights into their health benefits, medicaluses,andsafetyguidelines.
At the heart of this platform are powerful deep learning models like ResNet50 and VGG16, trained on large collections of plant images. In addition, the platform will feature a rich database of medicinal plants, offering users easy access to essential information such as active compounds, therapeutic purposes, and usage precautions all presented in a user-friendly and accessibleway.
1. L. D. S. Pacifico, L. F. S. Britto, and collaborators [14] emphasize the crucial role that medicinal plants play inhealthcare,whilealsoacknowledgingthechallenges in identifying them accurately due to the sheer number and diversity of species. They point out that many existing recognition systems lack the precision and adaptability needed for reliable identification. To tackle this issue, the team created a specialized dataset focused on capturing key leaf characteristics such as texture and color. They then developed an automated recognition system powered by five different machine learning algorithms. Their most successful model achieved over 97% accuracy, proving that carefully selected visual features can significantly boost identification performance. Their findings demonstrate the powerful synergy between traditional botanical knowledge and modern AI tools, with promising implications for both accurate identificationandconservationofmedicinalplants.
2. C. Sivaranjani, L. Kalinathan, R. Amutha, and their team [4] addressed the common problem of segmenting plant leaves in photos taken under varying lighting conditions. They introduced an innovative technique using the ExG-ExR index, which blends Excess Green and Excess Red components for

International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056
Volume: 12 Issue: 05 | May 2025 www.irjet.net p-ISSN:2395-0072
more reliable vegetation detection. With its zerothreshold approach, the method simplifies the segmentation process and maintains accuracy regardless of lighting differences. Once the leaf regions are isolated, color and texture features are extracted and analyzed using Logistic Regression, achieving an impressive accuracy of 93.3%. This streamlined preprocessing approach not only enhancesdataqualitybutalsosupportsmoreaccurate performance in deep learning models used for medicinalplantrecognition.
3. R. Janani and A. Gopal [12] explored the use of deep learning, specifically Convolutional Neural Networks (CNNs)andpretrainedmodelslikeVGG16andVGG19, to identify native Ayurvedic medicinal plants. Their research is especially impactful for healthcare professionals, pharmacies, public health organizations, and communities. It also has practical applications in remote areas where drones can captureplantimagesforidentificationpurposes.Their dataset includes leaf images from 64 medicinal plant species found in Kerala. The CNN model alone achievedanaccuracyof95.79%,whiletheVGG16and VGG19 pretrained models delivered even better results,with97.8%and 97.6%accuracy,respectively. These findings highlight the effectiveness of using pretrained deep learning models in identifying medicinalplants,especiallyinreal-worldapplications.
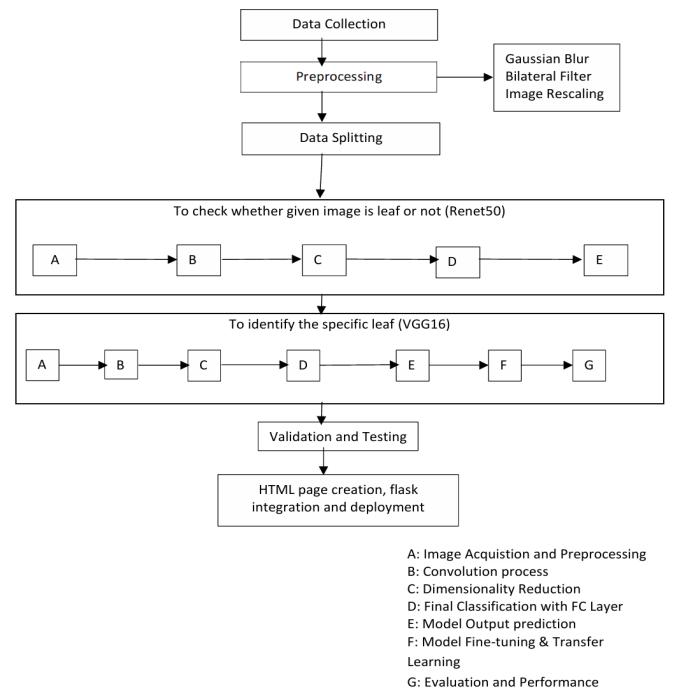
Dataset Collection & Exploratory Data Analysis (EDA)
For this project, we used the Indian Medicinal Leaves Dataset available on Kaggle. It features a wide variety of
commonlyusedAyurvedicplantspecies.Asafirststep,we performed Exploratory Data Analysis (EDA) to better understand the dataset looking into aspects like class distribution, image quality, and consistency in image resolution.Duringthisphase,wealsoidentifiedchallenges such as class imbalance and the subtle visual similarities betweensomeplantspecies.
Topreparetheimagesformodeltraining,weresizedallof them to 224×224 pixels to match the input size expected byConvolutionalNeuralNetworks(CNNs).Toimprovethe model's ability to generalize and reduce the risk of overfitting, we applied several data augmentation techniques, including image rotation, flipping, zooming, and brightness changes. In addition, pixel values were normalized to a 0–1 range to standardize input and improvemodelperformance.
Thedatasetwassplitintothreesubsets:70%fortraining, 15%forvalidation,and15%fortesting.Thissplitallowed us to train the models effectively while also testing their abilitytoperformwellonnew,unseendata.
Model-selection
WeevaluatedtwopopularpretrainedCNNarchitectures:
● ResNet50 – A deeper network with residual connections, well-suited for handling relatively smallerdatasetswhilemaintaininghighaccuracy
● VGG16 – A straightforward and deep model, known for its strength in fine-grained image classificationtasks.
Both models were fine-tuned by adjusting their final layers to match the number of medicinal plant classes in ourdataset.
Model training was carried out using categorical crossentropy as the loss function. We experimented with both the Adam and SGD optimizers. To prevent overfitting, we incorporated techniques like dropout and early stopping. Performance was evaluated using accuracy and other key metrics to ensure the models were both effective and reliable.
To make the system user-friendly, we created a simple webinterfaceusingHTML, allowinguserstouploadplant images and view identification results. The backend was built using Flask, which handled image processing and model predictions. Finally, the application was deployed onHeroku,makingiteasilyaccessibletothepublic.

Volume: 12 Issue: 05 | May 2025 www.irjet.net p-ISSN:2395-0072
ResNet50 is a powerful deep convolutional neural networkmadeupof50layers,andit'spartofthebroader Residual Network (ResNet) family. It was introduced by He et al. in their groundbreaking 2015 paper, "Deep Residual Learning for Image Recognition." What makes ResNet unique is its use of residual connections, also knownasskipconnections,whichallowthemodeltolearn differences (or "residuals") from previous layers rather than trying to learn entirely new features from scratch. This clever approach helps solve common problems in deep networks, such as vanishing gradients and performance degradation as networks get deeper. ResNet50 is often pre-trained on massive datasets like ImageNet, where it learns to recognize general visual features like edges, textures, and shapes. Thanks to this broad knowledge base, ResNet50 is highly effective for transfer learning making it a great fit for specialized tasks like medicinal plant identification, even when workingwithlimiteddata.
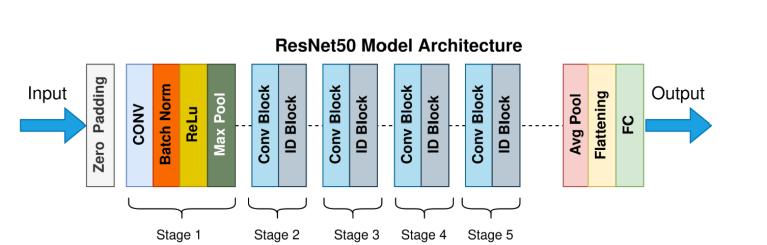
HowResNet50IsUsedforMedicinalPlantIdentification:
1.DatasetPreparation:
Thefirststepinvolvescollectingadiverseandhigh-quality image dataset of medicinal plant leaves, captured in variouslightingandenvironmentalconditions.Eachimage isresizedto224×224pixelsandconverted toRGBformat tomatchResNet50’sinputrequirements.
2.InputFormatting:
Tomaintainthespatialstructureoftheimageandprevent loss of detail around the edges, *zero-padding* is applied beforefeedingtheimageintothemodel.
3.InitialFeatureExtraction:
The image is passed through the first convolutional layer, whichdetectsbasicvisualelementslikeedgesandtexture. Batchnormalizationfollowstostabilizeandacceleratethe learningprocessbystandardizingtheactivationsfromthe previouslayer.
4.ActivationFunction(ReLU):
A ReLU (Rectified Linear Unit) function introduces nonlinearity, allowing the network to understand more complexpatternsinthedata.
5.Downsampling(MaxPooling):
A max-pooling layer helps shrink the size of the feature maps by keeping only the most important details, making the model more efficient without losing key information.This helps the model run faster and more efficiently.
6.DeepFeatureLearning:
The network continues with a series of convolutional and identity blocks. Convolutional blocks include filters, batch normalization, activation, and shortcut connections that help the network adjust as it learns. Identity blocks carry information directly across layers via skip connections, helping the model retain both new and old features as it learns.
7.OvercomingDeepNetworkChallenges:
These residual connections are vital they prevent problems like vanishing gradients and help the model learn better as it gets deeper. As the model progresses through more layers, it learns to pick up on finer details suchasuniqueleafshapes, patterns,andsurfacetextures, which are essential for distinguishing between similarlookingspecies.
8.LearningatMultipleLevels:
Skip connections allow the model to learn simple and complex features at the same time, which is especially important for identifying medicinal plants that may look verysimilartoeachother.
9.FeaturePoolingandClassification:
Once feature extraction is complete, average pooling compresses the data into a manageable format without losing valuable information. These features are then flattened into a vector and passed through a fully connectedlayer,wherethenetworkmapsthemtospecific plant categories. The final output uses a Softmax function to convert scores into probabilities, showing the most likelyplantspecies.
10.FinalPrediction:
In the last step, the model provides its best guess for the plant's identity. This prediction helps botanists, researchers, and everyday users correctly identify medicinal plants. It also supports broader efforts in International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056

International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056
Volume: 12 Issue: 05 | May 2025 www.irjet.net p-ISSN:2395-0072
conservation, education, and the documentation of herbal medicine.
VGG16 is a well-established deep learning model developedbytheVisualGeometryGroupattheUniversity ofOxford.Itwasintroducedinthelandmarkpaper*"Very Deep Convolutional Networks for Large-Scale Image Recognition."* This architecture includes 16 layers with learnable parameters 13 of which are convolutional layers, followed by 3 fully connected layers. A standout featureofVGG16isitsconsistentuseof3×3convolutional filters, which gives it a clean, straightforward structure that’s easy to understand and apply. Known for its strong performance in image classification, VGG16 is often used with pre-trained weights from the ImageNet dataset. These pre-trained weights help the model recognize commonvisualelementslikeedges,textures,andpatterns.
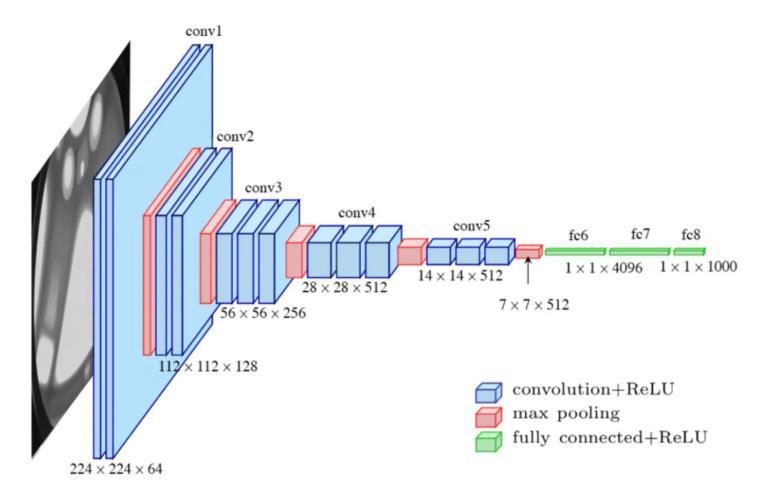
- 2: VGG16Architecture
HowVGG16HelpsinIdentifyingMedicinalPlants:
1.ImageCollectionandPreparation:
The process begins with gathering a varied and highquality dataset of medicinal plant images, captured under different lighting and environmental conditions. Each image is resized to 224×224 pixels with three color channels(RGB)tomatchthemodel’sinputrequirements.
2.ImageEnhancementandAugmentation:
Preprocessing techniques are applied to enhance image clarity this includes normalization and noise reduction. To further strengthen the model’s ability to generalize across different plant types and conditions, data augmentation techniques like rotation, flipping, and zooming are used. These artificially expand the dataset andhelpthemodeladapttoreal-worldvariations.
3.FeatureExtractionThroughConvolution:
The preprocessed images pass through a series of convolutional layers.Initially,the network identifies basic features such as edges and corners.As the image moves deeperintothenetwork,moreintricatedetailssuchasleaf shape, vein patterns, textures, and color differences are recognized. ReLU activation functions follow each layer, addingnon-linearitytohelpthenetworkcapturecomplex visualpatterns.
4.ReducingComplexitywithMax-Pooling:
To manage computational load and reduce the risk of overfitting, max-pooling layers are introduced after some convolutional blocks. These layers reduce the size of the feature maps while preserving important features, shrinking them from 224×224×64 to 7×7×512. This enhances the model's efficiency and helps it stay focused onthemostimportantfeatures.
5.TransitiontoClassification:
Theprocessedfeaturemapsaretransformedintoasingledimensional vector, making them ready for the classification stage.This vector is passed through a series of fully connected (dense) layers, where the model starts associatingpatternswithspecificplantspecies.
6.FullyConnectedLayersandCustomOutput:
Thedenselayersknownas fc6andfc7 eachcontain4096 units with ReLU activations, helping the network identify detailed traits unique to each plant. The final fully connected layer, originally set up to classify 1000 ImageNet classes, is customized to match the number of medicinalplantsinthedataset.
7.GeneratingPredictionswithSoftmax:
A Softmax function is applied to the final output, converting raw model outputs into probability scores for eachplantclass.Thishelpsthesystemmakeconfidentand interpretablepredictions.
8.LeveragingTransferLearning:
Toacceleratetrainingandimproveperformance,VGG16is initialized with weights learned from the ImageNet dataset. This allows the model to build on its existing knowledge of general visual patterns and tailor it to the specific task of medicinal plant identification. After training, its performance is evaluated using accuracy, precision, recall, and F1-score to ensure reliability. Once validated, the model can be deployed in user-friendly applications like web tools or mobile apps making medicinal plant recognition accessible to researchers, educators,andnatureenthusiasts.

International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056
Volume: 12 Issue: 05 | May 2025 www.irjet.net p-ISSN:2395-0072
A comparison was made between the pre-trained CNN models, RESNET50 and VGG16, and a simple CNN model. Thestudyusedleafimagesfrom64differentplantspecies, with a dataset of 64,000 images 1,000 samples per species. The data was broken down into 70% for training the model, 10% for validation, and the final 20% for testing its performance. The models were trained for 25 epochswitha batchsizeof 16,andthe final accuracywas measuredaftertesting.
As mentioned earlier, the first step was to build a basic convolutional neural network fromscratch,trainitonthe dataset,andassessitsperformance.TheCNNarchitecture consisted of three convolutional layers, each using a 3x3 kernel with ReLU activation, followed by 2D max-pooling layers of 2x2. The first three convolutional layers focused onfeatureextraction,andtheoutputwasthenpassedtoa fully connected dense layer for classification. In addition, the pre-trained VGG16 model, which had already learned from a vast and varied dataset, was employed to extract featuresandclassifytheimages.
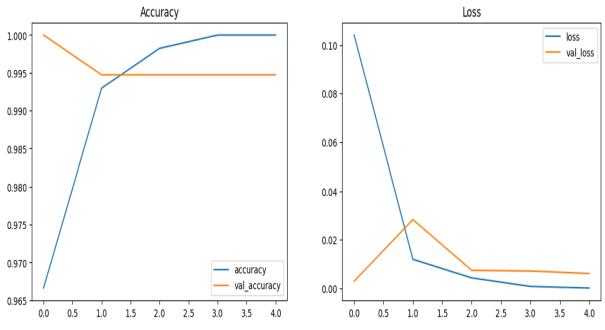
Fig - 3: AccuracyandLossGraphsofResNet50
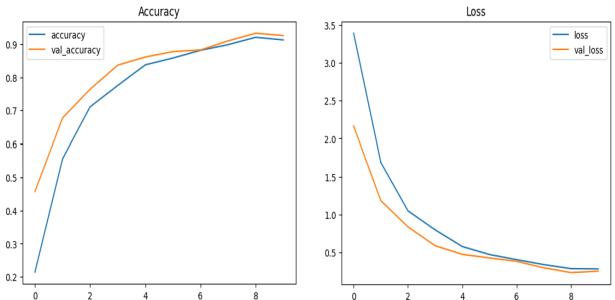
Fig - 4:AccuracyandLossGraphsofVGG16
The training loss values, training accuracy values, validation loss values, and validation accuracy values at successiveepochsofResNet50andVGG16aspresentedin Figures3and4respectively Theprogressofaccuracyand loss functions, during the training process and validation process,areshowninthegraphs.Thebluelineshowsthe trainingloss.Theorangelineshowstrainingaccuracy.The green line shows validation loss, and the red line shows validationaccuracy.
Table 1: Resnet50ClassificationReport
The above table ResNet50 classification report shows outstandingperformancewithanoverallaccuracyof99%. It achieved a precision of 0.99 and recall of 1.00 for leaf images, and perfect precision (1.00) with a recall of 0.99 for unknown classes. The high F1-scores and consistent macroandweightedaverageshighlightthemodel'sstrong balanceandreliabilityacrossthedataset.
The above table represents the VGG16 classification report. The model reached a total accuracy of 94%. The balanced macro and weighted averages (both at 0.94) indicate strong and consistent performance across the diversemedicinalplantdataset.

International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056
Volume: 12 Issue: 05 | May 2025 www.irjet.net
Table 3:Resultswithaccuracy

The table illustrates sample results of the medicinal plant detection system, showcasing correct identification of plants like Aloe Vera, Amla, Betel, and Eucalyptus along with their medicinal uses and achieved accuracy. This demonstrates the model's effectiveness in accurate classification and informative output for each plant species.
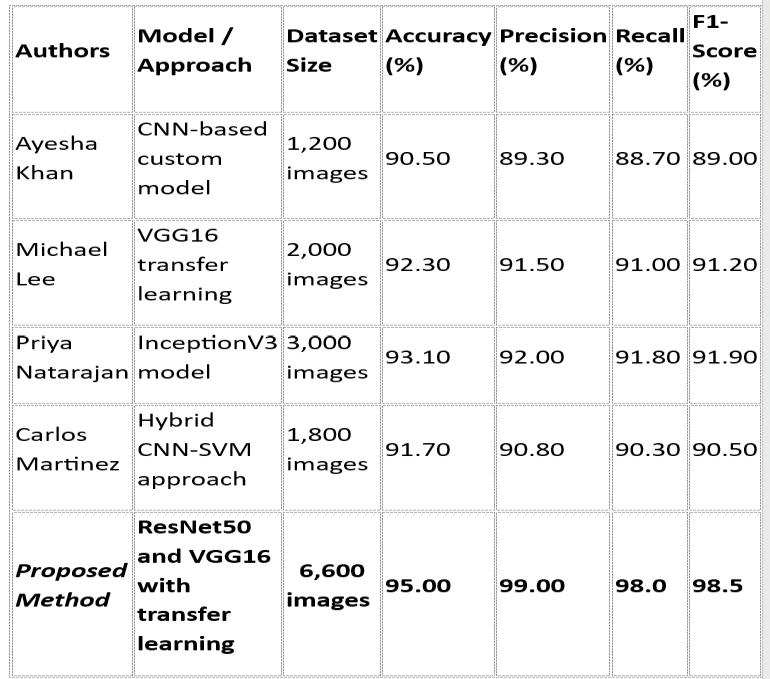
Table 4: Comparisiontabletootherpapers
p-ISSN:2395-0072
The table presents a comparative analysis of existing models with the proposed ResNet50 and VGG16 hybrid approach, which outperforms others by achieving 95% accuracyand98.5%F1-scoreusingalargerdataset.
The development of a medicinal plant detection system usingtheResNet50deeplearningmodeloffersanaccurate and efficient method for identifying medicinal plant species from images. By leveraging convolutional neural networks and transfer learning, the model effectively captures complex features such as leaf patterns, textures, and shapes. Preprocessing techniques like resizing and normalization ensured high-quality inputs, while skip connections and batch normalization helped maintain performance and mitigate challenges like vanishing gradients. The system provides a scalable, automated solution for botanists and researchers, enhancing plant identification and conservation efforts. Future improvements with larger datasets and further optimizationscanexpanditsapplicabilitytoawiderrange ofplantspecies.
The future of medicinal plant detection using deep learningholdsimmensepotentialacrossvariousdomains. Expanding the system with larger, globally sourced datasets covering a wider variety of plant species and diverseenvironmentalconditionswillgreatlyimprovethe model's accuracy and generalizability. Integration with mobile applications and IoT devices can facilitate realtime, field-based identification, making the technology accessible to farmers, botanists, and practitioners worldwide. Additionally, incorporating multi-modal data suchasgeographicallocation,soilcomposition,and climatic conditions can enhance the system's predictive capabilities, The incorporation of augmented reality (AR) can offer interactive identification experiences, enhancing educationaleffortsandsupportingfieldresearch.
Advancements in model optimization, including refined transferlearningtechniquesandtheadoptionofemerging architectureslikeVisionTransformers,canfurtherelevate performanceand efficiency. Beyondspeciesidentification, themodelcanbeextendedtopredictmedicinalproperties and detect plant diseases, increasing its utility in agriculture, pharmacology, and natural medicine. The system also offers valuable support for conservation efforts by monitoring rare and endangered medicinal plantsintheirnaturalhabitats.Finally,thedevelopmentof a comprehensive, AI-powered medicinal plant database can serve as a vital resource for researchers, educators, and conservationists, promoting sustainable use and preservationofplantbiodiversity

International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056
Volume: 12 Issue: 05 | May 2025 www.irjet.net
1. M.E.Rahmani,A.Amine,andM.R.Hamou,“Plant leaves classifica tion,” ALLDATA 2015, vol. 82, 2015.
2. Sabu,K.Sreekumar,andR.R.Nair,“Recognitionof ayurvedic medicinal plants from leaves: A computer vision approach,” in Image Information Processing (ICIIP), 2017 Fourth International Conferenceon.IEEE,2017,pp.1–5.
3. G.Agarwal,P.Belhumeur,S.Feiner,D.Jacobs,W.J. Kress, R. Ra mamoorthi, N. A. Bourg, N. Dixit, H. Ling, D. Mahajan et al., “First steps toward an electronic field guide for plants,” Taxon, vol. 55, no.3,pp.597–610,2006.
4. C. Sivaranjani, L. Kalinathan, R. Amutha, R. S. Kathavarayan and K. J. Jegadish Kumar, "RealTime Identification of Medicinal Plants using Machine Learning Techniques," 2019 International Conference on Computational Intelligence in Data Science (ICCIDS), Chennai, India, 2019, pp. 1-4, doi: 10.1109/ICCIDS.2019.8862126.
5. Dr. Ayesha Khan ,“A Lightweight CNN ArchitectureforMedicalImageClassification.”
6. Mallah, J. Cope, and J. Orwell, “Plant leaf classification using probabilistic integration of shape, texture and margin features,” Signal Processing, Pattern Recognition and Applications, vol.5,no.1,2013.
7. Prof.MichaelLee,“TransferLearningwithVGG16 forDiseaseDetectioninMedicalImaging.”
8. Sahay and M. Chen, “Leaf analysis for plant recognition,” inSoftwareEngineeringandService Science (ICSESS), 2016 7th IEEE International Conferenceon.IEEE,2016,pp.914–917.
9. G. Cerutti, L. Tougne, J. Mille, A. Vacavant, and D. Coquin,“Understandingleavesinnaturalimages–a model-based approach for tree species identification,” Computer Vision and Image Understanding, vol. 117, no. 10, pp. 1482–1501, 2013.
10. Y. Sun, Y. Liu, G. Wang, and H. Zhang, “Deep learning for plant identification in natural environment,” Computational intelligence and neuroscience,vol.2017,2017.
11. Sabu and K. Sreekumar, “Literature review of image features and classifiers used in leaf based plant recognition through image analysis
p-ISSN:2395-0072
approach,” in Inventive Communication and Computational Technologies (ICICCT), 2017 InternationalConferenceon.IEEE,2017,pp.145–149.
12. R. Janani and A. Gopal, "Identification of selected medicinal plant leaves using image features and ANN," 2013 International Conference on Advanced Electronic Systems (ICAES), Pilani, India, 2013, pp. 238-242, doi: 10.1109/ICAES.2013.6659400.
13. VenkataramanandN.Mangayarkarasi,“Computer vision based feature extraction of leaves for identification of medicinal values of plants,” in Computational Intelligence and Computing Research (ICCIC), 2016 IEEE International Conferenceon.IEEE,2016,pp.1–5.
14. L. D. Pacifico, V. Macario, and J. F. Oliveira, “Plant classification using artificial neural networks,” in 2018 International Joint Conference on Neural Networks(IJCNN).IEEE,2018,pp.1–6.
15. Transfer learning with fine-tuned deep CNN ResNet50 model for classifying COVID-19 from chestX-rayimages-ScienceDirect
16. Ben Rahman, Djarot Hindarto (PDF) Implementation of ResNet-50 on End-to-End ObjectDetection(DETR)onObjects
17. Mohit Chhabra;Rajneesh Kumar, “An Efficient ResNet-50basedIntelligent DeepLearningModel toPredictPneumoniafromMedicalImages.”
18. Jiahui Tao;Yuehan Gu;JiaZheng Sun;Yuxuan Bie;Hui Wang, “Research on vgg16 convolutional neural network feature classification algorithm basedonTransferLearning.”
19. K Kiran;Arpanpreet Kaur, Deep Learning-Based Plant Species Identification using VGG16 and Transfer Learning: Enhancing Accuracy in BiodiversityandAgriculturalApplications
20. Dr. Priya Natarajan , “Deep Feature Extraction UsingInceptionV3forMedicalDiagnosis.”
21. Dr.CarlosMartinez, “HybridCNN-SVMModelfor EnhancedMedicalImageClassification.”
22. T. M. Mitchell et al., “Machine learning. wcb,” 1997.
23. S. A. Dudani, “The distance-weighted k-nearestneighbor rule,” IEEE Transactions on Systems, Man, and Cybernetics, vol. 1, no. 4, pp. 325 327, 1976.

International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056
Volume: 12 Issue: 05 | May 2025 www.irjet.net p-ISSN:2395-0072
24. Criminisi, J. Shotton, and E. Konukoglu, “Decision forests for clas sification, regression, density estimation, manifold learning and semi supervised learning [internet],” Microsoft Research,2011.
25. E. Rumelhart, G. E. Hinton, and R. J. Williams, “Learning internal representations by error propagation,” California Univ San Diego La Jolla InstforCognitiveScience,Tech.Rep.,1985.
26. S. S. Haykin, Neural networks: a comprehensive foundation.TsinghuaUniversityPress,2001.
27. F.Pedregosa,G.Varoquaux,A.Gramfort,V.Michel, B.Thirion,O.Grisel,M.Blondel,P.Prettenhofer,R. Weiss, V. Dubourg, J. Vander plas, A. Passos, D. Cournapeau, M. Brucher, M. Perrot, and E. Duch esnay, “Scikit-learn: Machine learning in Python,” JournalofMachineLearningResearch,vol.12,pp. 2825–2830,2011.
28. L.Buitinck,G.Louppe,M.Blondel,F.Pedregosa,A. Mueller, O. Grisel, V. Niculae, P. Prettenhofer, A. Gramfort, J. Grobler, R. Layton, J. Van derPlas, A. Joly, B. Holt, and G. Varoquaux, “API design for machine learning software: experiences from the scikit-learn project,” in ECML PKDD Workshop: LanguagesforDataMiningandMachineLearning, 2013,pp.108–122.
29. F.Cholletetal.,“Keras:The python deeplearning library,”AstrophysicsSourceCodeLibrary,2018.
30. M. Friedman, “The use of ranks to avoid the assumptionofnormalityimplicitintheanalysisof variance,” Journal of the american statistical association,vol.32,no.200,pp.675–701,1937.
31. P. Nemenyi, “Distribution-free multiple comparisons,” in Biometrics, vol. 18, no. 2. INTERNATIONALBIOMETRICSOC1441IST,NW, SUITE700,WASHINGTON,DC20005-2210,1962, p.263.