
Volume:12Issue:05|May 2025 www.irjet.net
International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056 p-ISSN:2395-0072



Volume:12Issue:05|May 2025 www.irjet.net
International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056 p-ISSN:2395-0072


M. Sri Varsha Shwetha1 , K. Aparna2 , R. Kirthiga 3 , A. Mahalakshmi 4
1 Assistant Professor, Department of Information Technology, Meenakshi College of Engineering, Chennai, Tamil Nadu, India
2-4Students, Department of Information Technology, Meenakshi College of Engineering, Chennai, Tamil Nadu, India
Abstract -An Early detection of congenital heart defects (CHDs)isessentialtoallowprenatalcareandmedicaltreatment in time. This system is a machine learning-based system used for fetal heart assessment from ultrasound images. The system works on an end-to-end basis, processing images through various stages, including preprocessing, feature selection, and classification, to separate normal from abnormal fetal heart images. The workflow begins with image enhancement techniques that are used for preprocessing to maintain uniform conditions across the images. Then, K-means clustering is implemented to automatically cluster the images into normal and abnormal categories. To assist in classification, features are extracted with the help of two complementary approaches: Histogram of Orientated Gradients (HOG), which highlights the shape and structural patterns of the heart, and Local Binary Patterns (LBP), which emphasize the texture details. Finally, these features are fed to an XGBoost classifier to distinguish between the classes. This proposed approach thus allows clinicians to reach fast yet reliable decisions, which in turn ensures better prenatal outcomes and early treatment planning ofcongenitalheartdiseases.
Keywords: Fetalheartdetection,Congenitalheart defects (CHDs), Ultrasound imaging, Machine learning, Image preprocessing, K-means clustering, Feature extraction, Histogram of Oriented Gradients (HOG), Local Binary Patterns (LBP), XGBoost classifier, Prenatal diagnosis, Medicalimageanalysis.
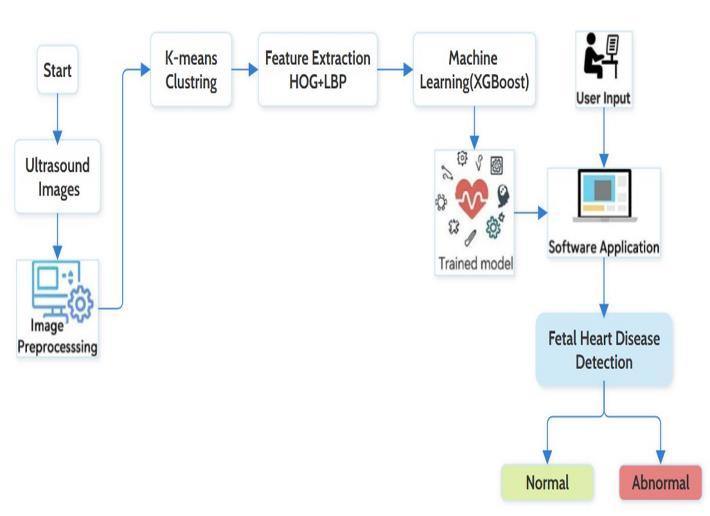
Congenital heart defects (CHDs) constitute one of the leading reasons for neonatal complications. Their early diagnosis is relevant for timely intervention and appropriateprenataltreatment.Inthisproject,wepresent a machine-learning-based method for fetal heart assessmentthroughultrasoundimages.Thestageissetby preprocessing, which entails improving image clarity and standardizingimagequality.Thecronies,K-means,cluster the images into normal and abnormal patterns by visual pattern matching. Then, the extraction of features takes place: HOG captures shape information and LBP texturerelated information from each image. These features are combined to form a unique signature for each image that
can be used to feed into an XGBoost classifier. This model trains itself on the differences between a healthy and abnormal fetal heart with increased accuracy. Reducing subjectivityfromthediagnosisprocesswillallowforearly intervention, thereby improving the condition of mother andchild.
Theexistingsystemforheartdiseasepredictionutilizes traditional machine learning methods such as Support Vector Machine (SVM), Random Forests (RF), and logistic regression (LR) on structured clinical data [1][5][8]. Such systemsworkondatasetsliketheClevelandHeartDisease Dataset (CHDD) and a private heart disease dataset amalgamated from various medical centres [4][6]. These systems, however, require manual engineering of features from parameters within tables such as age, sex, cholesterol, blood pressure, and various other clinical indicators. Such features are picked on the basis of expert knowledge and this lack of consistency generally lowers the level of performance of the model and its generalizability across populations [7] [9]. Moreover, the existing systems do not take into account ultrasound images, thereby losing out on critical spatial, shape, or texture features crucial for a diagnosis [2][3][10]. Such limitations are pivotal for complicated diagnoses that rest onvisualpatterns,particularlyforanearly-stagedetection process. This severely limits, in turn, the capability of traditionalmodelsforearlydetectionanddiagnosis.
This work proposes an intelligent fetal heart detection system that uses a machine learning pipeline. It functions through combined shape and texture analysis to classify fetal heart ultrasound images into normal and abnormal. using K-means clustering. Features are extracted using Histogram of Oriented Gradients (HOG) for shape and Local Binary Pattern (LBP) for texture. Then, the XGBoost classifier uses the extracted features to detect the fetal heart condition The resultant system aims at minimizing manual interpretation errors while also reinforcing earlystage diagnosis accuracies These predictions and alerts

Volume:12Issue:05|May 2025 www.irjet.net
International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056 p-ISSN:2395-0072

are available to be disseminated through a connected platform.
Ultrasound Image → Image Preprocessing → K-Means Clustering → Feature Extraction (HOG + LBP) → XGBoost Classifier→Result
Anotheroptionwouldbetolinkawebinterfacethat tracksclassificationresultsandalertsaboutanydetected abnormalconditions.
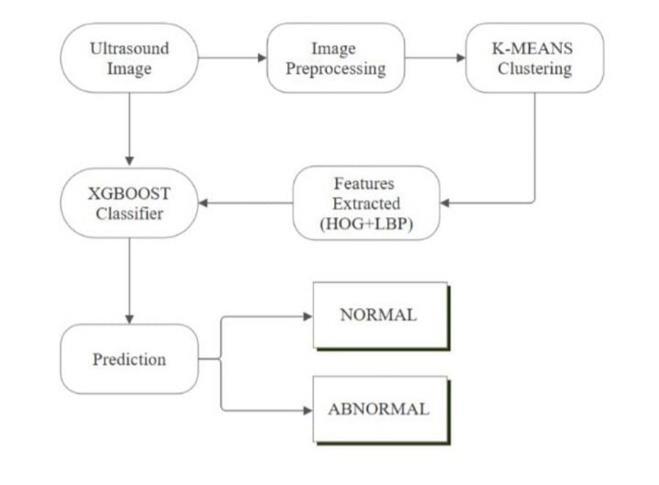
4.1. Data Preprocessing:
In ultrasound imaging, the role of the Preprocessing module is particularly important in the analysis phase. Ultrasoundrawimagesaresometimesafflictedwithnoise, low contrast, and other forms of degradation which can impedetheperformanceofanyanalyticalprocedure.This module implements image denoising, contrast improvement, and normalization in order to make the images clearer and more consistent throughout the dataset. Preprocessing, through standardizing the quality ofimages,ensuresthatthedatasuppliedtotheclustering andclassificationstepsisreliable.Enhancingtheaccuracy of the fetus heart detection system depends on the effective and reliable feature extraction, which in great part,dependsonthelevelofpreprocessingperformed.

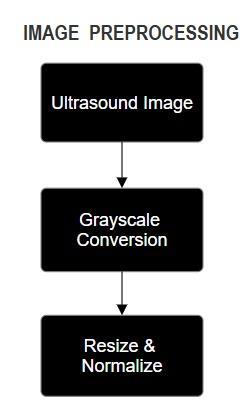
4.2.
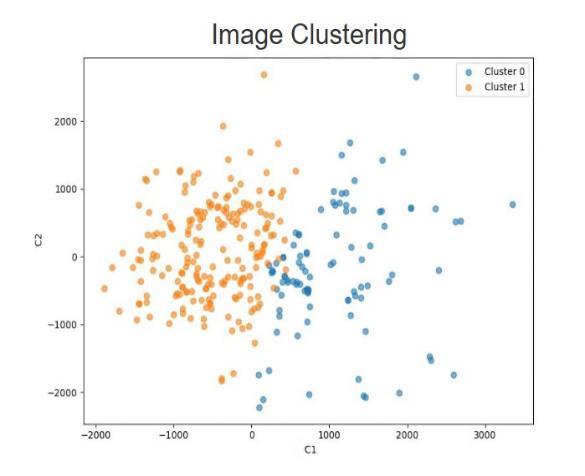
The Image Clustering Module employs K-means clustering to classify ultrasound images into normal and abnormal categories. This method classifies images based ontheirvisualfeaturesbygroupingcommonpatternslike pixels with similar intensity. For normal and abnormal patterns of fetal heart structures, clustering mitigates manual annotation and organizes images for feature extraction and classification. This serves to streamline data preparatory processes, increasing complexity while improving the reliability and detection accuracy of the system.
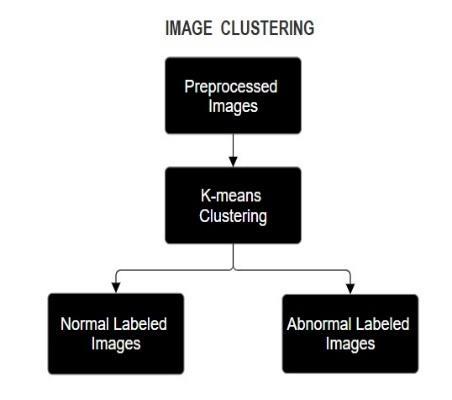
4.3.
The Feature Extraction Unit identifies and pulls out meaningful characteristics from preprocessed ultrasound images to support classification. These features are typically based on shape and texture, which are essential in distinguishing between normal and abnormal fetal heart conditions. To extract shape-related features, the Histogram of Oriented Gradients (HOG) method is used, while Local Binary Patterns (LBP) are employed for capturing texture patterns. Together, these features are combinedintoacomprehensivefeaturevectorthatserves as a unique representation of each image. This vector is then passed to the classification model, which uses it to determine the fetal heart status. the fetal heart detection system.

International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056 p-ISSN:2395-0072

Volume:12Issue:05|May 2025 www.irjet.net
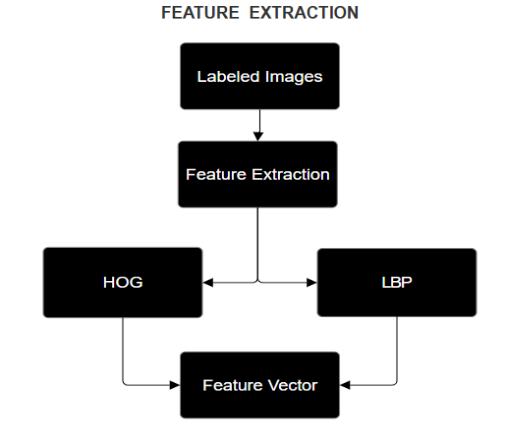
FeatureExtractionFlowchart
4.4. Classification
The Classification Module takes the feature vectors generated by the Feature Extraction Module and places them as Normal or Abnormal in the fetal heart images. It is the center of the ML pipeline. The extracted features are classified by using powerful classifiers, such as XGBoost-widely regarded as the topmost classificationalgorithmintermsof accuracy andefficiency-andtheresultantclasslabelisassigned to the image. The classifier was trained on a labeled data set of ultrasound images so as to be able to correctlydiscriminatebetweennormalandabnormal heart cases. Once the classification of an image is done by the system, the result is output so that systems can be used in timely decisions by the cliniciansaboutfetalhealth.
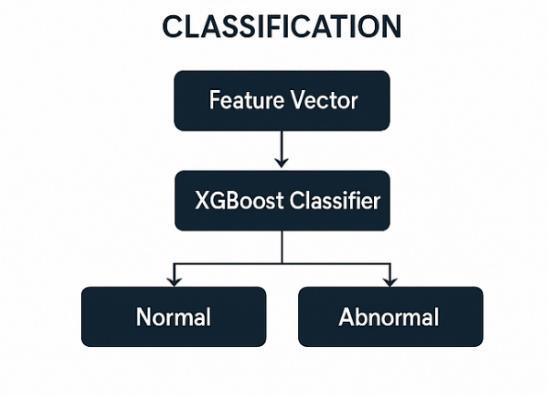
Fig-5: ClassificationFlowchart
4.5. Evaluation &Testing
In order to determine how well the proposed systemperformsonfreshdata,itwasevaluatedwith ultrasound images not included in the training set. Thisstepverifiesthatthemodelisnotoverfittingand memorizing training data instead of learning the underlying patterns. A confusion matrix was generated to assess the classification performance of the system towards the fetal heart images by determining the correct and abnormal classification.

In addition, classical evaluation measures: accuracy, precision, recall, and F1-score were computed to provide a descriptive analysis of the system’s evaluation results.Inaddition,all ofthe misclassified cases were analyzed in detail to find underlying reasons in the model which may aid to enhance the system.
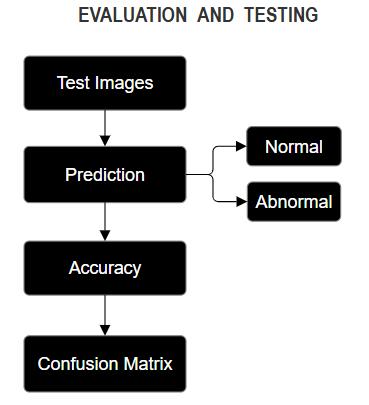
Fig-6: Evaluation&Testing Flowchart
5. SOFTWARE APP
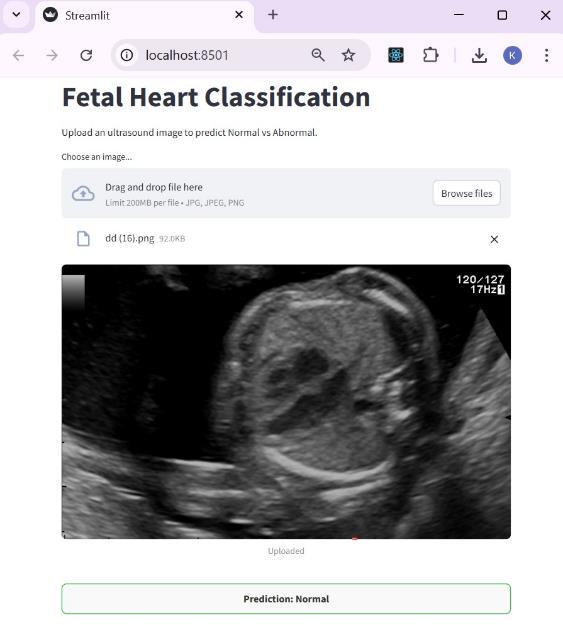
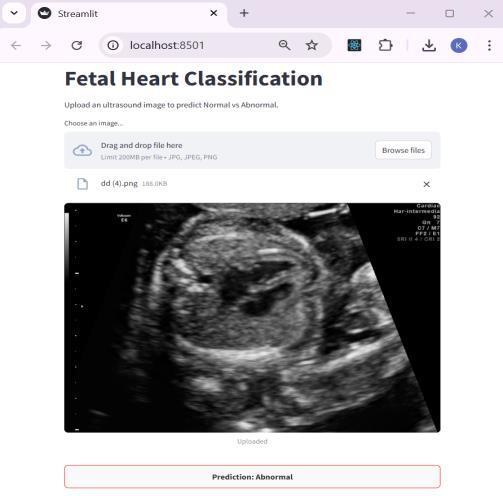
The application was developed with Streamlit, whichisknownasa webframework forPython.Ithas an intuitive interface which allows users to upload a fetal heart ultrasound image and obtain a prediction resultalmostimmediatelyNormalorAbnormal.Dueto its simple design, the app is classified as lightweight and easy to use. It is browser-based, meaning that it can be accessed easily during consultations, making it effortless to use during clinical assessments, even withoutextensivetechnicaltraining.
The system supports a straight forward interface where users can submit fetal heart ultrasound images for analysis. Users can surf through images and select one;thesystemwillthenprocesstheimageandissuea ‘normal’ or ‘abnormal’ diagnosis. This interface strives to optimize ease of use. The UI permits diagnostics to be performed quickly and accurately in a clinical setting,andstreamlinedworkflows.
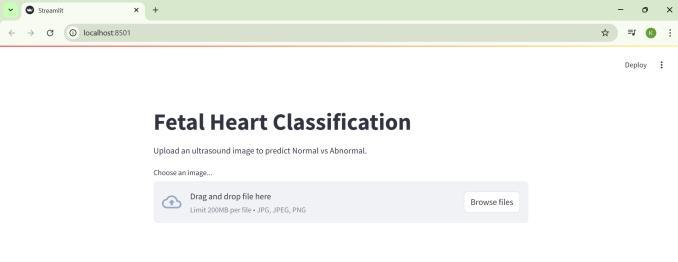
Fig-7: UserInterface

International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056 p-ISSN:2395-0072

Volume:12Issue:05|May 2025 www.irjet.net
6.1. Image Preprocessing
Input:Rawultrasoundimageoffetalheart
Process:
Loadtheultrasoundimages from the data set
Converttheimagesintograyscale
Resizeintoapredeterminedsize(64*64)
Normalizethepixelsbyscalingfrom0to1
K-means clustering-based detection of fetalheartregion
Output: Prepared ultrasound image for the next stageofsegmentation.
6.2. Image Segmentation using K-means Clustering
Input:Preprocessedultrasoundimage.
Process:
ApplyHOGtogetedgeandshapefeatures
ApplyLBPtogettexturefeatures
Convert both HOG and LBP into number form(vectors)
Combinethemintoonefeatureset
Save features with their label (Normal or Abnormal)
Output:LabeledimagesasNormalandAbnormal
6.3. Feature extraction
Input:LabeledimagesasNormalandAbnormal
6.3.1 Shape Feature Extraction Using HOG (Histogram ofOriented Gradients)
Process:
Divides the image into small cells, usually 8*8pixels.
Calculate gradients and orientation of pixelswithinacell.
Create histogram of gradients orientation foreachcell.

Normalize the histograms over large spatial areas, known as blocks, comprising Several cells.
Output: HOG descriptors that describe the shape featuresofthefetalheartregion
6.3.2 Texture Feature Extraction Using LBP (Local Binary Patterns)
Process:
Compare the intensity values for the central pixelsofaneighborhood,usually3*3or5*5.
Generate a binary code from these intensity comparisons.
Convertthebinarycodeintoadecimalnumber thatrecordstheLBPhistogram.
Output: Texture features (LBP descriptors) of the fetalheartregion.
6.4. CreatingFeatureVector
Input: Shape Features (HOG) and Textures Features(LBP)
Process:
Integrate HOG and LBP features into one vector.
Output: One feature vector that integrates both shapeandtextureinformationofthefetalheart.
6.5. Classification Using XGBoost
Input:Featurevector.
Process:
Train an XGBoost Classifier with labelled dataset (Normal/Abnormal images and their correspondingfeaturevectors).
Use the trained XGBoost model on the input image'sfeaturevector.
Classification will provide the prediction label(NormalorAbnormal)
Output:Classificationresult:NormalorAbnormal.

International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056 p-ISSN:2395-0072

Volume:12Issue:05|May 2025 www.irjet.net
A machine vision-based fetal heart condition detectionsystemisunderdevelopmentforthisresearch. Thesystemstartswithimageacquisitionandprocessing to ensure quality, that is, noise reduction, contrast enhancement, etc. The system then comprises image grouping via K-means clustering, where images are placed into either "Normal'' or ''Abnormal'' categories depending on their visual patterns. In this proceeding, features are extracted; shape features and texture-based HOG and LBP features are extracted. Then an XGBoost classifierisusedtoclassifytheheartconditionasNormal and Abnormal. The intelligent system helps in early detectionofsmallercomplicationstherebyimprovingthe accuracy of diagnosis and aiding the physician in deciding faster and make more reliable decisions for betterprenatalcareandoutcomes.
Withtheproposedmachinelearningsystem,a94%accurate classification of fetal heart conditions from ultrasound images has been realized. It showed a good generalization ability when subjected to test images beyond those it was able to train on, having rightly classified 47 out of 50 images and incorrectly classifying three. Other key performance metrics further strengthen the robustness of the model, with a precision of 95%, recall of 93%, and F1-score of 94%. From the detection standpoint,thispresentsabalancebetweenfalse-positive rates and true-positive rates for abnormal occurrence identification. The whole scheme with K-means clustering, HOG,LBP feature extraction,andclassification through XGBoost tiltsthescaletowardsthestronger side of the system. Thus, acting as a prompt, reliable, and objective tool for the early diagnosis of congenital heart diseases, particularly when expert opinions are absent withintheclinicalsetting.



International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056 p-ISSN:2395-0072

Volume:12Issue:05|May 2025 www.irjet.net
9. CONCLUSION
This study highlights several key concepts in the ultrasound images: image preprocessing, detection of the fetal heart, feature extraction with HOG and LBP, and classification through XGBoost with 94% accuracy. The system very reliably detects the fetal heart condition as either normal or abnormal so that it reduces much of the manual interpretation and now allows faster and consistent diagnosis. Such automation can be a boon for many countries where an expert is scarce. Having strong performance metrics and an adaptable design behind it, the system assists clinicians in the early diagnosis of congenital abnormalities to ensure better prenatal care andbringhealthyoutcomesformotherandchild.
10. FUTURE ENHANCEMENTS
1. Real-TimeIntegration
Facilitate an interface with ultrasound machines so that inline analysis can be performed during the scans.
2. Mobile app creation
Visionthedevelopmentofamobileapplicationthat is less resource-intensive and can allow fetal heart detectioninfar-offplaces
3. Diagnosis with MoreThan OneClass
Improve the model further so that several specific types of congenital heart defects can be diagnosed insteadofnormalvs.abnormal.
11. ACKNOWLEDGEMENT
As Information Technology students, our engagement in this project has been nothing short of beneficial. Participating on the design of a system which detects fetal heartbeats based on machine learning

principlesfacilitatedsome ofthetheoretical knowledge to be put into practice in an experimental setting. The successful completion of this project has sharpened our technical skills and taught us how to work together on solvingactualproblemsinlife.
[1] H. F. El-Sofany, “Predicting heart diseases using machine learning and different data classification techniques,” IEEE Access, vol. 12, pp. 106146-106160, Aug.2024,doi:10.1109/ACCESS.2024.3437181.
[2] T. V. Sushma, N. Sriraam, P. M. Arakeri, and S. Suresh, “Classification of fetal heart ultrasound images for the detection of CHD,” in Advances in Electrical and ComputerTechnologies,Springer,2021,pp.443-452,doi: 10.1007/978-981-15-9651-3_41.
[3] L. Yan, S. Ling, R. Mao, H. Xi, and F. Wang, “A deep learningframeworkforidentifyingandsegmentingthree vessels in fetal heart ultrasound images,” Bio Medical Engineering Online, vol. 23, no. 1, 2024, doi: 10.1186/s12938-024-01230-
[4]S.AlrabieandA.Barnawi,“HeartWave:Amulticlass dataset of heart sounds for cardiovascular diseases detection,” IEEE Access, vol. 11, pp. 118722-118736, 2023,doi:10.1109/ACCESS.2023.3325749.
[5] B. Ramesh and K. Lakshmanna, “A novel early detection and prevention of coronary heart disease framework using hybrid deeplearning model and neural fuzzy inference system,” IEEE Access, vol. 12, pp.2668326695,2024,doi:10.1109/ACCESS.2024.336653 7.
[6] A. Gudigar et al., “An Automated System for the DetectionofHeartAnomaliesUsingPhonocardiograms:A Systematic Review,” IEEE Access,12:138399–138428,2024. https://doi.org/10.1109/ACCESS.2024.3465511
[7] H. Khan et al., “Heart Disease Prediction Using Novel Ensemble and Blending Based Cardiovascular DiseaseDetectionNetworks:EnsCVDD-NetandBlCVDDNet,” IEEE Access, 12: 109230–109254, 2024. doi:10.1109/ACCESS.2024.3421241
[8] M. S. A. Reshan et al., “A Robust Heart Disease PredictionSystemUsingHybridDeepNeuralNetworks,” IEEE Access, 11: 121574–121591, 2023. doi:10.1109/ACCESS.2023.3328909
[9] W. Sun et al., “Machine Learning Based Prediction of Cardiovascular Diseases,” IECE Transactions on Internet of Things, 2(2): 50–54, May 2024. doi:10.62762/TIOT.2024.128976

Volume:12Issue:05|May 2025 www.irjet.net
International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056 p-ISSN:2395-0072

[10] P. Saha et al., “Self-Supervised Normality Learningand DivergenceVectorGuidedModel Merging for Zero-Shot Congenital Heart Disease Detection in Fetal Ultrasound Videos,” arXiv:2503.07799, submitted Mar.2025.
