The first HiFi benchtop system bringing industry-leading and exceptionally accurate long reads to your lab.

Effortless HiFi workflow from sample prep to results


















6 EIGHT BABIES BORN FOLLOWING MITOCHONDRIAL DONATION
The birth of eight babies with a greatly reduced risk of developing mitochondrial DNA disease provides hope for those waiting to access mitochondrial donation.
15 NEW COELIAC DISEASE TEST REMOVES NEED FOR GLUTEN CONSUMPTION
The test can detect coeliac disease with up to 90% sensitivity and 97% specificity — even in patients following a strict gluten-free diet.
16 NOVEL DRUG CANDIDATES FOR NERVE PAIN AND ISCHEMIC DISEASE
The drugs’ discovery paves the way for potential new treatments for health conditions linked to tissue stress and inflammation, without accompanying side effects.
21 BIOMATERIAL HELPS REVERSE AGING IN THE HEART
The material could open the door to therapies that rejuvenate the heart by changing its cellular environment, rather than the heart cells themselves.
24 BUDGET CUTS PUT RESEARCH INTEGRITY UNDER STRESS
As respected institutions face cuts to their funding, there is a question mark over both the volume and quality of research that can be conducted.
26 GENETIC ADAPTATION TO COLD-WATER DIVING IN KOREA’S HAENYEO
The Haenyeo, from the Korean island of Jeju, are renowned for their ability to dive in frigid waters without the aid of breathing equipment — even while pregnant.
28 REAL-TIME SEQUENCING HELPS COMBAT GOLDEN STAPH
Tracking bacterial changes during serious golden staph infection can give clinicians immediate insights to personalise treatment.
30 PRECISE MEASUREMENT OF RADIOACTIVITY IN TINY SAMPLES
The new method offers a streamlined approach, identifying radioactive elements and quantifying their level of radioactivity in even tiny samples.
32 TINY ANTIBODIES PROTECT AGAINST HENIPAVIRUSES, CORONAVIRUSES
Researchers have been investigating the potential of tiny antibodies, sourced from the family of land animals known as camelids, to protect against certain viruses.

Last issue, I stated my intention to attend NATA’s Accreditation Matters conference in late July. I can now confirm that I did attend the event, the conference sessions were indeed fascinating, and several were also very relevant to issues that are currently in the spotlight — namely forever chemicals (also known as perand polyfluoroalkyl substances, or PFAS) and forensic science.
Steve Beaman from the NSW Environmental Protection Agency (EPA) noted at the event that PFAS are ubiquitous in the environment because they are very resistant to disintegration — this being the reason they were so widely used in manufacturing in the first place. Here in Australia, efforts are now in place to minimise exposure to PFAS, with PFOS, PFOA and PFHxS now banned from local manufacture, import and use, and the National Health and Medical Research Council (NHMRC) recently releasing revised drinking water guidelines with lower recommended PFAS exposure levels.
But as various jurisdictions seek to regulate, restrict and ban PFAS, Daniel Slee from the National Measurement Institute said there should be serious discussion about industries where their use is essential (indeed, the Australian ban makes an exemption for PFAS used for medical devices, medicines and biologicals). Furthermore, Dr Paul Byleveld from NSW Health said that if we continue to lower recommended exposure levels, there’ll
be pressure on labs to be able to measure to those lower levels, which may not be feasible for them.
One area where labs are expected to perform to high standards is DNA testing, particularly when it comes to forensic science — which is why it was so concerning when, just days after the conference, Forensic Science Queensland (FSQ) was back in the headlines following a long-awaited review that highlighted the need for significant reform. In particularly worrying news, the review found that FSQ has a current median turnaround time for DNA results in major crimes of 412 days — which would be bad enough even if the extra time taken was for quality assurance purposes, but it turns out the review also identified environmental contamination within the laboratory as well as sample contamination, on top of a toxic, reactive workplace culture.
So how do you foster a quality culture in your laboratory? When it comes to forensic science, Dean Catoggio of the Australia New Zealand Policing Advisory Agency (ANZPAA) recommends consulting the recently developed Principles of Authentic Quality Culture in Forensic Science Service Provision, which outline four key areas that enhance service quality and foster continuous improvement. NATA’s Claire Lillee meanwhile suggests seeking NATA accreditation, which serves as a formal recognition that your organisation meets certain standards — and that it is committed to continuous improvement, transparency and fixing any mistakes that do occur. And Dekel Faruhi from Proofig AI, in his article on page 24 of this magazine, recommends the use of quality-
assurance tools to safeguard research integrity and maintain trust in your lab.
Some of our other articles this issue have a focus on improving patient safety. For example, the story on page 15 describes a landmark blood test that can identify coeliac disease even when a patient is on a gluten-free diet, unlike current diagnostic tests that require regular gluten consumption in order to be effective. The article on page 29 meanwhile reveals how real-time tracking of bacterial changes during serious Staphylococcus aureus (golden staph) infection gives patients the best chance of recovery, by avoiding unnecessary treatments, minimising side effects and limiting antibiotic resistance. And in a massive milestone, our lead story on page 6 delves into the birth of eight babies in the UK following mitochondrial donation, which limits the transmission of disease-causing mitochondrial DNA mutations from mother to child — and should be available in Australia before too long.
Happy reading!


Recently, it has become possible to identify brain biomarkers associated with Alzheimer’s disease through a blood sample, which should in future offer a cost-effective method for identifying those at greatest risk of developing Alzheimer’s disease and prioritising them for preventive treatments. But until now, brain biomarkers associated with Alzheimer’s disease have mainly been studied in older individuals. A new study, published in The Lancet Healthy Longevity, provides insights into biomarker levels and associated factors starting from middle age, years or even decades before any decline in cognitive functions becomes apparent.
As part of the Cardiovascular Risk in Young Finns Study, conducted at the University of Turku, biomarkers associated with Alzheimer’s disease were measured from blood samples of middle-aged participants (aged 41–56) and their parents (aged 59–90), with a total sample size of 2051 individuals. A key finding was that a high biomarker concentration in the parent, particularly the mother, may be associated with higher biomarker levels in the middle-aged offspring.
The researchers additionally found that kidney disease may be linked to higher levels of biomarkers in middle age. The APOE ε4 gene, which increases the risk of Alzheimer’s disease, was meanwhile associated with higher blood-based biomarker levels in older age, but not yet in middle age.
The researchers emphasised that it is not yet possible to definitively diagnose Alzheimer’s disease with a blood sample, as the method is still limited by the lack of well-known reference values. Additionally, it remains unclear which confounding factors influence biomarker concentrations in blood related to Alzheimer’s disease. Therefore, the interpretations of the biomarkers obtained from blood sample could lead to misdiagnosis.
“In order to reliably use blood-based biomarkers for Alzheimer’s disease diagnosis in the future, more research is needed across different population and age groups to standardise reference values,” concluded study leader Suvi Rovio.
Specialised Therapeutics (ST) has announced the Australian registration of its drug SKYTROFA (lonapegsomatropin) as a once-weekly injectable therapy for paediatric growth hormone deficiency (GHD). The Therapeutic Goods Administration (TGA) says it has approved SKYTROFA as a treatment for growth failure in children and adolescents aged from 3–18 years due to insufficient endogenous growth hormone secretion.
GHD is a rare disease affecting 2–3/10,000 Australians, with approximately 2000 children thought to be living with the condition. It occurs when the pituitary gland, located at the base of the brain, does not produce an adequate level of growth hormone, which is essential for promoting healthy growth in children. GHD is typically characterised by slow height growth, resulting in short stature, and may also include delayed puberty, impaired hair growth and headaches. The goal of treatment is to restore normal levels of growth hormone in the body, reduce physical symptoms, enhance metabolic health and improve quality of life.
SKYTROFA is a prodrug of somatropin administered once weekly, designed to provide sustained release of active, unmodified somatropin. This allows the medicine to slowly release unmodified growth hormone into the body over the course of the week, removing the need for daily injections. The starting dose depends on the patient’s body weight and is then adjusted individually by the treating endocrinologist based on the patient’s response to treatment.
SKYTROFA is being registered in Australia under an exclusive distribution agreement with biopharmaceutical company Ascendis Pharma — whose TransCon technology platform was used to develop the treatment — that covers Australia, New Zealand, Singapore, Malaysia, Brunei, Thailand and Vietnam. The TGA registration was based on the results of Ascendis Pharma’s three pivotal Phase 3 clinical trials, which collectively treated more than 300 paediatric patients diagnosed with GHD and found the drug was generally well tolerated.
“We are delighted to have secured TGA registration of SKYTROFA for eligible Australian children with growth hormone deficiency,” said ST CEO Carlo Montagner. “Beyond short stature, children and adolescents with the condition may experience considerable physical and psychosocial impacts on their daily life, such as poor concentration, decreased strength or muscle development, fatigue, and reduced quality of life.
“This announcement also represents a significant milestone for Specialised Therapeutics, marking the second endocrinology therapy we have successfully registered in Australia.”

Lauren Davis
Eight children in the UK have been born following a pioneering licensed IVF technique to reduce the risk of mitochondrial diseases, known as mitochondrial donation. The UK was the first country to approve laws to allow the use of this groundbreaking technology in 2015, followed by Australia in 2022.
Mitochondria produce the energy required for life and contain a small piece of DNA that only encodes some of the instructions required for energy production. Harmful mutations in mitochondrial DNA can result in reduced availability of energy, particularly affecting tissues that have high energy demands (such as the heart, muscle and brain). Mitochondrial DNA is maternally inherited, meaning mitochondrial diseases are passed from mother to child (males can be affected but do not pass on the disease), and despite years of research there is still no cure. Attention has therefore turned to IVFbased technologies to reduce the risk of disease by limiting transmission of disease-causing mitochondrial DNA mutations from mother to child — with so-called ‘pronuclear transfer’ (PNT) showing particular promise.



PNT is performed after the egg is fertilised. It involves transplanting the nuclear genome (which contains all the genes essential for our individual characteristics, such as hair colour and height) from an egg carrying a mitochondrial DNA mutation to an egg donated by an unaffected woman that has had its nuclear genome removed. The resulting embryo inherits its parents’ nuclear DNA, but the mitochondrial DNA is inherited predominantly from the donated egg.
PNT was pioneered in human eggs over almost two decades by a team based at Newcastle University and the Newcastle upon Tyne Hospitals NHS Foundation Trust, led by Professor Mary Herbert. It is now being offered to prospective parents through a research study, as part of an integrated program that also includes preimplantation genetic testing (PGT) — a procedure that helps couples avoid passing on genetic conditions by testing their embryos for such conditions. In accordance with regulations, PNT is offered only to those women who are unlikely to benefit from PGT treatment.
The team’s work to date has been published across two papers in The New England Journal of Medicine (NEJM)
The reproductive outcomes paper
At the time of reporting, clinical pregnancies were confirmed in eight of 22 (36%) patients who underwent PNT as part of the study and 16 of 39 (41%) of patients who underwent PGT. PNT has so far resulted in eight births to seven women (including one set of identical twins) and one further pregnancy, while PGT has resulted in 18 births.
Despite being born to mothers at high risk of transmitting serious disease caused by mutations in mitochondrial DNA, levels of disease-causing mitochondrial DNA mutations in the PNT newborns’ blood were either undetectable or present at low levels. According to the researchers, the presence of such mutations results from carryover of maternal mitochondria surrounding the nuclear DNA at the time of transplantation; a known limitation of mitochondrial donation technologies.
“The findings give grounds for optimism,” said Herbert, who was lead author of the reproductive outcomes paper. “However, research to better understand the limitations of mitochondrial donation technologies will be essential to further improve treatment outcomes.
“Mitochondrial donation technologies are currently regarded as risk-reduction treatments owing to carryover of maternal mitochondrial DNA during the mitochondrial donation procedure. Our ongoing research seeks to bridge the gap between risk reduction and prevention of mitochondrial DNA disease by addressing this problem.”
The clinical outcomes paper
All eight PNT babies were healthy at birth (whether born by normal vaginal delivery or elective caesarean section), had normal weight for their gestational age, and were described as developing normally and meeting relevant milestones. The team noted that three babies overcame separate early health issues that they were not able to attribute directly to mitochondrial
donation; these included some brief startles, which resolved without treatment; high blood fats (which also affected the mother during pregnancy) and cardiac arrhythmia (treated with a low-fat diet and anti-arrhythmic medication, respectively); and a urinary tract infection that responded quickly to antibiotic treatment.
The level of disease-causing mitochondrial DNA mutation was measured in blood and urine cells and was undetectable in five babies. Three babies had low levels of disease-causing mitochondrial DNA mutations — 5 and 9%, 12 and 13%, and 16 and 20% in blood and urine respectively. These levels are well below the 80% level required for clinical disease for these mutations, suggesting they would not be responsible for any of the children’s health conditions. At follow-up at 18 months, the level of the disease-causing mutation in the child with 5 and 9% was undetectable in blood and urine. The team will continue to offer assessments up to the age of five years, with the aim of detecting any patterns in childhood conditions.
“While longer term follow-up of children born following mitochondrial donation is of paramount importance, these early results are very encouraging,” said Bobby McFarland, Director of the NHS Highly Specialised Service for Rare Mitochondrial Disorders (Newcastle Hospitals NHS Foundation Trust) and Professor of Paediatric Mitochondrial Medicine at Newcastle University, who serves as first author on the clinical outcomes paper.
“We believe the follow-up process we have put in place is thorough, since it allows us to detect and review even minor health conditions
All eight PNT babies were healthy at birth (whether born by normal vaginal delivery or elective caesarean section), had normal weight for their gestational age, and were described as developing normally and meeting relevant milestones.
in children born after pronuclear transfer, such as a urinary tract infection.”
What does this mean for Australia?
Herbert relocated to Melbourne’s Monash University in 2023 to help establish mitochondrial donation in Australia, following the passage of Maeve’s Law the previous year, meaning Australians will be able to benefit directly from what has been learnt in the UK. That same year, the Medical Research Future Fund (MRFF) provided $15 million for the Monash-hosted mitoHOPE Program, to pilot the introduction of mitochondrial donation into Australian clinical practice.
Since late 2023, the Monash team and their partners have been working with the National Health and Medical Research Council (NHMRC) to gain a licence to train embryologists in mitochondrial donation, as a first step towards a clinical trial and clinical application of mitochondrial donation. According to Professor John Carroll, Director of the Monash Biomedicine Discovery Institute and Head of mitoHOPE, the
UK studies should give Australia momentum to follow suit.
“We hope to soon obtain our first licence from the NHMRC’s Embryo Research Licensing Committee, so that we can begin training our IVF embryologists in the procedure using donated eggs,” Carroll said.
“The birth of eight babies with a greatly reduced risk of developing mitochondrial DNA disease provides hope for those waiting to access mitochondrial donation and shines new light on the path toward introducing mitochondrial donation in Australia. The mitoHOPE team aims to start recruiting into the clinical trial in about a year, but this is dependent on us gaining the necessary licences.”
Herbert said the UK results were a long time in the making, and that she hopes the successful outcomes will help in navigating Australia’s “rather complex regulatory system”.
“I look forward to continuing this work at Monash University and I hope that the new treatment can soon become available to Australian families affected by these devastating diseases,” she concluded.

A major phase III clinical trial, led by Swedish researchers, has shown that a significantly shorter course of radiotherapy for localised prostate cancer is just as safe and effective as the traditional eightweek schedule — even 10 years after treatment. The trial’s findings, presented at ESTRO 2025, give both patients and doctors greater confidence in choosing this short-course approach, dubbed ‘ultra hypo-fractionated radiotherapy’.
For many prostate cancer patients, radiotherapy is a standard treatment option that offers outcomes comparable to surgery, particularly for localised disease, while allowing men to maintain much of their daily routine during treatment. However, traditional radiotherapy schedules typically span several weeks, which can be burdensome for patients and put pressure on healthcare structures and radiotherapy capacity.
Seeking to address this, the HYPO-RT-PC clinical trial enrolled 1200 men with intermediate- to high-risk localised prostate cancer. Participants were randomly assigned to receive either short-course radiotherapy — 42.7 Gray (Gy) delivered in seven sessions over 2.5 weeks — or standard-course radiotherapy (78 Gy delivered in 39 sessions over eight weeks). The researchers assessed survival, cancer recurrence and treatment-related side effects, including urinary and bowel symptoms.
The researchers found that delivering precision radiotherapy over 2.5 weeks is equally successful in beating prostate cancer as the standard eight-week approach, with both options producing similar disease control rates and survival a decade after treatment. Failure-free survival (no return of cancer or need for additional treatment) was 72% in the short-course group vs 65% in the standard group; overall survival was 81% for short-course vs 79% for standard; prostate cancer-specific mortality was 4% in both groups; and urinary and bowel symptoms were similar in both groups (mostly mild to moderate).
“These long-term findings confirm previous five-year results from the trial, showing that delivering fewer, higher doses over a shorter period works just as well as the standard approach — not just in theory, but in real-world clinical practice,” concluded Associate Professors Per Nilsson and Adalsteinn Gunnlaugsson, who led the 10-year outcome analysis of the HYPO-RT-PC trial at Skåne University Hospital and Lund University.
“For patients, this means less disruption to daily life and potentially lower healthcare costs — without compromising outcomes and safety.”
Researchers at The University of Queensland (UQ) have developed a new cell cultivation technique that uses algae — and it’s said to be cheaper, greener and more ethical than current methods. Their work has been published in Biotechnology Journal
Dr Melanie Oey and her team at UQ’s Institute for Molecular Bioscience combined a new type of Queensland algae, Chlorella sp. BDH-1, with mammalian cells to improve the process of growing tissue cells. Oey said the BDH-1 algae was chosen because it does not consume glucose and doesn’t compete with mammalian cells for food; indeed, it naturally creates a more supportive environment for muscle cells to thrive.
“In the human body blood delivers oxygen and removes waste, but in a lab setting you don’t have that system,” Oey said.
“By adding algae we’re essentially creating a mini-symbiosis or mutually beneficial interaction where the algae provide oxygen and take away waste, helping the cells grow better.”
The research could benefit tissue engineering and regenerative medicine, such as growing new skin for burns victims, with accelerated growth of 3D tissues and skin grafts. In cell cultures the researchers reported a more than 80% increase in cell growth, up to three times the number of usable cells, and cell cultures that remained viable for longer. There was also a 50% reduction in the need to use animal cells.
“The algae act like tiny life-support systems that can solve multiple problems at once,” Oey said.
“Our work shows muscle cells co-cultivated with the algae grow faster, live longer and require fewer expensive additives.”
The method also has the potential to benefit other applications, such as growing cultivated meat more affordably. Food Standards Australia New Zealand recently approved the sale of lab-grown or cell-cultured meat — created by growing or multiplying individual animal cells — but cost remains a barrier.
“Growing meat in the lab is expensive largely due to the nutrients and oxygen the cells need and the waste they produce,” Oey explained.
“Our research could make cultivated meat a sustainable, affordable, ethically acceptable alternative protein source.”
Other applications for the algae in cell culture include growing organoids to use in testing drugs, thus avoiding the need for animal testing; and growing and testing cells more efficiently as part of pharmaceutical manufacturing, with lower overheads and fewer inputs.


Researchers say they have found the genes linked to obsessive compulsive disorder (OCD), after identifying 30 regions on the human genome associated with this debilitating yet often misunderstood mental health condition. Their study, which involved over 200 investigators around the world, has been published in the journal Nature Genetics
OCD is often categorised as an obsession with cleaning or checking. However, it can take many forms, including worrying about harming others, doubts about relationships, existential worries, inappropriate sexual thoughts or persistent concern about responsibilities.
“People who suffer from OCD are driven by a fear they’ve done something wrong, or that they’re going to do something wrong,” explained clinical psychologist and OCD specialist Dr Emily O’Leary.
“They worry they could hurt their loved ones or themselves. For people with OCD, it’s like being trapped in their own worst nightmare.”
The new study is the culmination of more than 20 years of sample collection. It involved more than 50,000 people with OCD and 2 million people who did not have OCD, and pinpointed approximately 250 genes linked to OCD. These genes were found to be most active in three key brain areas — the hippocampus, the striatum and the cerebral cortex — which aligns with previous neuroscience studies.
“This is the first study where we found actual genes that play a role in OCD, which is really exciting,” said Professor Eske Derks, senior group leader of the Translational Neurogenomics Laboratory at QIMR Berghofer, who co-supervised the study. “We’ve been working on this for many years, but these findings have dramatically increased our knowledge of the genetic basis of OCD.
“We found a really large number of genes which are shared with anxiety disorders, depression and also with anorexia nervosa. There’s a lot of overlap in the genes that cause these different mental health conditions.”
According to Derks, the next steps are to use these genetic discoveries to identify existing drugs that may be effective for OCD patients, which could pave the way for more innovative treatment options. O’Leary added that the findings offer an opportunity to speed up diagnosis and treatment.
“What happens in clinical practice is that clients will come to us typically 10–14 years after diagnosis, and by this time they are really unwell,” she said. “What this research shows us is that we can be more proactive and actually start looking at who is more likely to develop the disorder.”
Researchers at the Hebrew University of Jerusalem say they have developed a remarkably precise method to estimate a person’s age based on just a small DNA sample — with potential applications in medicine, aging research and forensic investigations.
Using cutting-edge artificial intelligence, the scientists created a tool called MAgeNet that can determine a person’s chronological age (the number of years since birth) with a margin of error as small as 1.36 years for individuals under 50 — and all it takes is a simple blood draw.
“It turns out that the passage of time leaves measurable marks on our DNA,” said Professor Tommy Kaplan, co-supervisor on the project. “Our model decodes those marks with astonishing precision.”
The secret lies in how our DNA changes over time through a process called methylation — the chemical ‘tagging’ of DNA by methyl group (CH3). By zooming in on just two key regions of the human genome, the team was able to read these changes at the level of individual molecules, then use deep learning to translate them into accurate age predictions.
The team’s study, published in the journal Cell Reports, analysed blood samples from over 300 healthy people, as well as data from a decade-long longitudinal analysis of the Jerusalem Perinatal Study (JPS). Their results show that the model worked consistently across a range of variables — like smoking, body weight, sex, and even different signs of biological aging.

“This gives us a new window into how aging works at the cellular level,” said co-supervisor Professor Yuval Dor. “It’s a powerful example of what happens when biology meets AI.”
The research also uncovered new patterns in how DNA changes over time, suggesting our cells encode age both randomly and in coordinated bursts — like ticking biological clocks.
“It’s not just about knowing your age,” said co-supervisor Professor Ruth Shemer. “It’s about understanding how your cells keeps track of time, molecule by molecule.”
The research could reshape how we approach health, aging and identity in the future. From helping doctors tailor treatments based on a person’s true biological timeline, to helping forensic investigators estimate the age of a suspect, the work deepens our understanding of how aging works —bringing us one step closer to decoding the body’s internal clock.

Scientists from the Centenary Institute and The University of Sydney have identified a previously unknown gateway into human cells — a receptor called AAVR2 — that gene therapy viruses can use to deliver therapeutic genes. Described in the journal Cell, the pathway could allow lower doses of virus to be used to treat a range of serious genetic disorders, including Duchenne muscular dystrophy, Pompe disease and haemophilia.
Gene therapies typically use modified viruses, known as adeno-associated viruses (AAVs), to deliver healthy genes into the body. These treatments have the potential to be life-changing for patients, their families and caregivers. However, they frequently require high vector doses to achieve therapeutic effects which in some cases can trigger severe immune responses, leading to serious complications or even death.
“We found that certain AAV types can use this newly identified receptor, AAVR2, to enter cells, providing an alternative to the previously known entry route,” said lead author Dr Bijay Dhungel, a researcher at the Centenary Institute’s Centre for Rare Diseases & Gene Therapy and The University of Sydney.
“Modulating this pathway can potentially make gene therapies safer, cheaper and more precise.”
Using advanced genetic, biochemistry and molecular biology techniques, the researchers showed that AAVR2 plays a crucial role in helping several AAV types enter cells more efficiently.
“We not only identified this new receptor AAVR2 but also discovered how it binds to the viruses that deliver the genes,” said co-senior author Dr Charles (Chuck) Bailey, Head of the Centre for Rare Diseases & Gene Therapy and a researcher at The University of Sydney.
“We then went a step further and engineered a miniature version of the receptor and demonstrated that this significantly enhances how efficiently the gene therapy is taken up in human cells and tissues. We believe this knowledge will ultimately improve the accessibility of gene therapies to patients.”
The researchers say their findings have important implications for the future of gene therapy, offering new strategies to tailor treatments, lower required doses and potentially avoid immunerelated complications that have limited some current approaches. The discovery also advances scientific understanding of how therapeutic viruses interact with human cells, which should assist development of the next generation of safe, effective and precision-guided gene therapies.



Many researchers are looking for an easy way to reduce their carbon footprint, use less plastic and be more sustainable around the lab. The Axygen HybridRack, from Corning, is designed to enable just that, marrying functionality and sustainability by pairing a polypropylene deck with a paperboard base to allow for more sustainability in science — without sacrificing quality or comfort.
The product’s innovative paperboard and plastic hybrid design is said to use 70% less plastic than a traditional tip rack. This hybrid design creates a stable platform allowing end users to pipette directly from the paperboard base — even with a multi-channel instrument. The paperboard base can be easily recycled using most kerbside recycling programs, which should reduce the amount of plastic being thrown away in the lab.
Corning’s hybrid design is sterile, non-pyrogenic, RNase/DNase-free, and human gDNA- and PCR-inhibition-free. It is available with a polyethylene filter and available with Axygen Maxymum Recovery surface for maximum sample recovery. The product is made in the USA in an ISO 13485 and ISO 14001 registered facility.
Pacific Laboratory Products www.pacificlab.com.au
PacBio’s Vega benchtop system empowers scientists to manage sequencing projects in-house, removing batching bottlenecks and unpredictable project timelines by putting the user in full control from start to finish.
Powered by HiFi sequencing, the system is designed to consistently deliver accuracy of 99.9% or higher per run. The delivery of long reads with high precision should give researchers the confidence to explore complex biological questions.
Each run on the system includes real-time analysis of DNA methylation status using advanced deep learning algorithms. With integrated primary analysis, barcode demultiplexing and automatic conversion to BAM format, users will get fast access to meaningful data directly off the instrument.
The product encodes sequence data, quality scores and methylation information at just 0.5 bits per base, which should make it up to 20 times more efficient than other long-read platforms. Less storage facilitates faster analysis and more streamlined workflows.

While Protein A and G remain the most common choices for antibody capture, their broad and unspecific Fc binding and potential for leaching from beads can compromise specificity and lead to interfering background in sensitive assays. Optimised for targeted precision and clean results, Proteintech’s VHH-based beads are designed to redefine specificity and purity in immunoprecipitation, offering species-specific IgG binding and no Protein A or G contamination.
The company’s anti-rabbit/anti-mouse VHH beads are conjugated with a mix of three high-affinity VHHs (Nanobodies) targeting rabbit IgG, mouse IgG1, and mouse kappa light chains. This combination enables broad compatibility with the most commonly used rabbit and mouse antibodies.
Whether the user is performing routine IPs across various antibody subtypes or working with antibodies of unknown isotype, the beads offer a powerful alternative to Protein A or Protein G, delivering clean, efficient pulldowns with consistent performance.
United Bioresearch Products Pty Ltd www.unitedbioresearch.com.au

The system enables researchers to generate reference-grade microbial genomes, resolve challenging repeat expansions, characterise full-length isoforms and assemble phased human genomes — all from a compact benchtop system. It has the ability to transform the lab into a centre of high-resolution genomic exploration, elevating sequencing projects and making HiFi sequencing more accessible. Bio-Strategy - Part of DKSH Group www.bio-strategy.com

The South Australian Genomics Centre (SAGC) has become the first certified service provider in Asia–Pacific for Parse Biosciences’ Evercode technology for single-cell sequencing. Evercode allows researchers to analyse gene expression from millions of individual cells in a single experiment, going far beyond the limits of traditional single-cell workflows.
The SAGC is pairing Evercode with its MGI DNBSEQ-T7 sequencer, claimed to be the fastest and most-efficient sequencing platform in Australia, to deliver a highly scalable, cost-effective pipeline maximising single-cell research. According to SAGC Centre Manager Dr Sen Wang, access to this technology opens up a diverse range of possibilities for researchers locally, nationally and abroad.
“This technology is transforming what’s possible,” Wang said. “By combining Parse’s scalable single-cell kits with our ultrahighthroughput sequencing capacity, we can now support projects of virtually any size and budget.”
Single-cell RNA sequencing (scRNA-seq) enables researchers to understand the unique gene expression profile of each cell within a complex sample, revealing insights into cell type, state, function and disease signatures. Traditional droplet-based methods can be limited by scale and cost, but Parse’s combinatorial barcoding approach enables researchers to fix and pool samples and run anywhere from a few thousand cells up to five million cells per run — achieving a tenfold reduction in cost.
“It simplifies sample handling, allows batching of fixed cells, and gives researchers much more flexibility, especially for large-scale or multi-timepoint studies,” said SAGC Genomics Innovation Lead Dr Munir Iqbal.
SAGC has already supported projects using this platform from leading institutions across Australia and New Zealand, including SAHMRI (where the centre is based), The University of Queensland, the University of Otago, The University of Melbourne, the Garvan Institute of Medical Research and the Menzies Institute for Medical Research.
A recent example includes a study led by Dr Stephen Blake and Professor David Lynn at SAHMRI and Flinders University, evaluating the immune response to therapeutic mRNA vaccines in immune cold colon cancer. Their research leveraged SAGC’s certified Parse workflow to batch and process fixed cells from multiple sources and timepoints — generating high-quality single-cell data for in-depth comparison.
“The Parse workflow was simple on our end and made it possible to fix and preserve cells collected at different timepoints for later processing by SAGC,” Blake said.
“Munir from SAGC was very knowledgeable in the workflow and did a great job with our samples, keeping us informed at each stage of this process.”
As demand grows, SAGC is expanding its team and services to keep pace. The certification also complements SAGC’s recent recognition as a Xenium Certified Provider for spatial transcriptomics — making it one of only two centres in Australia certified for this advanced spatial biology platform.
“By bringing together these technologies, SAGC has become a national centre of excellence for single-cell and spatial genomics,” Wang said.
“We’re giving researchers the tools to interrogate biology at an unprecedented level — one cell at a time.”
Accurate particle size analysis is essential across a wide range of industries — from pharmaceuticals and food to mining and battery manufacturing. But real-world conditions can often complicate laser diffraction measurements: bubbles, dust and other contaminants can interfere with scattering signals, leading to misleading particle size distributions.
The Mastersizer 3000+ from Malvern Panalytical addresses these challenges with features designed to help users get precise results every time, regardless of their experience level. With a dynamic range from 10 nm up to several millimetres in a single optical set-up, the product eliminates the need for hardware changes or complex reconfiguration. It’s fully compliant with ISO 13320 standards and can measure multimodal size distributions for both wet and dry sample analysis, delivering reproducibility and resolution even in the sub-micron range.

Built-in expert guidance and intuitive software interfaces empower users of all skill levels to perform complex measurements with confidence. Adaptive Diffraction is Malvern Panalytical’s patent-pending, machine-learning-based technology that recognises and isolates interference from environmental noise. With a high-speed acquisition rate of 10 kHz, the system identifies and separates transient scattering events (caused by bubbles or contaminants) from steady state sample data, for more reproducible particle size distribution. Size Sure uses Adaptive Diffraction to store sub-run measurement data and then applies machine-learning classification to gain a clear view of steady-state sample results vs transient events, which reduces the time spent troubleshooting inconsistencies.
For even deeper insight, the optional Hydro Insight accessory delivers real-time imaging of dispersed particles — offering visual confirmation of particle shape, size and dispersion quality. This is a useful tool for applications where morphology matters.
The Mastersizer 3000+ provides a smart, fast and robust way to get meaningful particle size data — helping scientists and engineers make well-informed decisions every time.
ATA Scientific Pty Ltd www.atascientific.com.au

HEMCO’s Vented Safety Enclosure (VSE) is offered in 61, 91 and 122 cm widths to accommodate an analytical balance and other small-scale lab processes.
Constructed with clear anodised aluminium and 1/4″ thick clear polycarbonate on all sides, the product has an efficient airflow design with airfoil and bypass, directing contaminates to baffled exhaust, and providing good airflow and containment performance for user protection.
The viewing sash is angled 15° for ease of viewing comfort with a 20 cm reach-in opening height. The sash swings up to provide full frontal access. The ergonomic design provides added user comfort. HEMCO Corporation www.hemcocorp.com
The AION is a gentle 3D live-cell confocal imager from Confocal NL, providing high-contrast images from thicker specimens such as organoids, tissue samples, and plant and animal model organisms. The system allows a 25 mm field of view (FOV) with a motorised pinhole, designed to provide spot-on confocality every time. The gentle conditions for live samples enable use in long-time-lapse experiments and imaging of low-signal samples with >96% QE. This makes the product particularly suitable for live-cell imaging.
The combination of sCMOS camera-based detection and a set of filters allows acquisition of images in both visible (VIS) and near-infrared (NIR). This is designed to allow improved temporal resolution, high sensitivity and a good signal-to-noise ratio, resulting in fast imaging with clean results. The additional motorised single-band emission filters result in >50 fps @ 3Kx3K or >1000 fps in fixed line mode (camera limited).
The system uses a line illumination through a slit pinhole, instead of a point illumination found in point scanner confocal microscopes. This provides high temporal resolution while reducing phototoxicity. Since phototoxicity is one of the biggest challenges for researchers, this makes AION suitable for live-cell imaging.
The product can be added to any widefield microscope, which will turn it into an advanced and fast-scanning confocal imaging system. Three models are available depending on the user’s needs: AION α, AION λ and AION Σ SciTech Pty Ltd www.scitech.com.au


Current diagnostic tests for coeliac disease require people to eat large amounts of gluten — the thing that makes them sick — for weeks in order to get an accurate diagnosis. Now, researchers at the Walter and Eliza Hall Institute (WEHI) and Novoviah Pharmaceuticals have developed a landmark blood test that can identify the condition in patients — even when they’re on gluten-free diets.
Coeliac disease is one of the most common autoimmune illnesses in Australia, caused by an immune reaction to the gluten protein found in wheat, rye and barley. While early diagnosis is critical to minimising long-term complications of the disease, up to 80% of cases around the world remain undiagnosed.
The diagnostic process can be confusing, as the reduction in symptoms from a gluten-free diet is not always a marker of coeliac disease. Furthermore, current coeliac testing methods do not work reliably for those on a gluten-free diet, and require regular gluten consumption to be effective. As a result, many people are deterred from seeking a definite diagnosis because they do not want to consume gluten and feel sick.
“There are likely millions of people around the world living with undiagnosed coeliac disease
simply because the path to diagnosis is difficult and, at times, debilitating,” said Associate Professor Jason Tye-Din, Head of WEHI’s Coeliac Research Lab and a gastroenterologist at The Royal Melbourne Hospital.
In 2019, WEHI researchers, working with Dr Robert Anderson (who went on to co-found Novoviah Pharmaceuticals), discovered that the immune marker interleukin 2 (IL-2) spiked in the bloodstream of people with coeliac disease shortly after they ate gluten. But could this critical signal still appear, even if no gluten had been consumed? Building on a promising pilot study in 2021, and using a new clinical diagnostic system developed with Novoviah Pharmaceuticals, the research team set out to test the performance and understand the science of an ‘in-tube’ gluten challenge blood test.
In their new study, published in the journal Gastroenterology , the researchers used blood samples from 181 volunteers recruited via The Royal Melbourne Hospital. This included 75 people with treated coeliac disease (on a glutenfree diet); 13 people with active, untreated coeliac disease; 32 people with non-coeliac gluten sensitivity; and 61 healthy controls. Participant blood samples were then mixed with gluten in a test tube for a day to see if the IL-2 signal appeared.
PhD researcher Olivia Moscatelli, who was diagnosed with coeliac disease at 18, said the team was thrilled to find the test could detect the condition with up to 90% sensitivity and 97% specificity — even in patients following a strict gluten-free diet.
“This breakthrough is deeply personal as it could spare others from the gruelling diagnostic process I had to endure,” Moscatelli said.
“Knowing I’ve played a role in this achievement is a powerful, full-circle moment.”
As seen in the previous study, the IL-2 signal only increased in the volunteers with coeliac disease, demonstrating the immune response to gluten can be detected in a tube, without a gluten challenge. The test thus represents a promising new tool to support diagnosis, Moscatelli said — especially for people who can’t be diagnosed with the currently available methods.
“We also found the strength of the IL-2 signal correlated with the severity of a patient’s symptoms, allowing us to predict how severely a person with coeliac disease might react to gluten, without them actually having to eat it,” she added.
Moscatelli noted that some diagnostic tests give false positives in the presence of other autoimmune diseases, such as type 1 diabetes or Hashimoto’s thyroiditis, but this was not an issue.
“This is largely because the technology we use is highly sensitive and can detect the IL-2 signal at exceptionally low levels,” she said. “It’s like the equivalent of being able to detect a single grain of sand in a swimming pool.”
While the ultrasensitive cytokine testing technology used for this study isn’t currently utilised in pathology labs, the researchers hope this will change in the future, which would allow the blood test to be widely used by healthcare professionals. The WEHI team is now collaborating with Novoviah Pharmaceuticals to confirm the test’s accuracy across diverse populations and accrue real-world data.
“This test could be a game changer, sparing thousands of people the emotional and physical toll of returning to gluten,” Tye-Din said. “It’s a major step towards faster, safer diagnosis.”

Australian and Swedish scientists have discovered novel drug candidates which could ultimately lead to new effective treatments for conditions caused by tissue stress and inflammation, including neuropathic pain and ischemia-reperfusion injury. Their work has been published in the journal PNAS
Neuropathic pain, also known as nerve pain, is often linked to inflammation in the peripheral and central nervous systems. Ischemiareperfusion injury is tissue damage that occurs when blood flow is restored to an area that has been deprived of oxygen, ie, following a heart attack.
The study, led by the Monash Institute of Pharmaceutical Sciences (MIPS) in collaboration with Uppsala University, focused on the discovery of drug-like candidates to target the adenosine A1 receptor (A1R) subtype.
Above: Cryo-EM structure of the human adenosine A1 receptor (A1R, white) in complex with the orthosteric agonist adenosine (magenta) and the allosteric modulator MIPS521 (cyan).
A1Rs are widely distributed in the brain and heart and play a key role in communication between neurons.
Health conditions triggered by tissue stress benefit from A1R activation, and thus this receptor has been identified as a promising target for ischemiareperfusion injury and chronic neuropathic pain. However, the successful development of drugs targeting A1R has remained challenging due to unwanted effects: both on-target, such as slowing heart rate, and off-target, caused by interactions with other adenosine receptor subtypes.
The research team used a combination of advanced technologies to discover a novel set of subtype-selective A1R positive allosteric modulators (PAMs) — drug candidates that specifically target A1R and enhance its activity without affecting other adenosine receptors. Unlike traditional agonists that fully activate A1R and often cause side effects like slowed heart rate, these PAMs act like a ‘dimmer switch’ rather than an on/off switch — subtly enhancing the receptor’s response only when and where it is naturally active, offering more precise control with fewer on-target effects.
The team’s discovery paves the way for potential new treatments for neuropathic pain, ischemia-reperfusion injury and other diseases linked to
tissue stress and inflammation, without accompanying side effects. Their work thus marks a pivotal advance in A1R-targeted drug development, according to co-first author Dr Anh Nguyen from MIPS.
“When the new drug candidates we’ve discovered bind to A1R they are able to modulate neuron activity in such a way that unwanted side effects, such as cardiac reactions, are no longer a hindrance,” Nguyen said. “This has been a significant hurdle in the development of drugs targeting A1R, so we are very excited by this discovery and its potential to enable safer, more effective treatments for a range of conditions.”
A1R is part of the G-protein-coupled receptor (GPCR) family — the largest drug target class behind many major life-changing medical advancements in recent decades. Drugs that target the GPCR family make up about 34% of all US FDA-approved drugs, including Cobenfy (for schizophrenia) and semaglutide (Ozempic, Wegovy) for diabetes and obesity.
Co-lead author Dr Lauren May, also from MIPS, said the team’s discovery can largely be attributed to significant developments in drug screening technologies and computational techniques.
“A much deeper pharmacological understanding of GPCRs, combined with technological advancements, has collectively transformed the potential of this incredible family of drug targets which, in recent years, has led to new classes of life-changing drugs for millions of people around the world,” May said.
“In particular, the study showcases the efficacy of using cryo-electron microscopy (cryo-EM) — a technology which enabled the team to identify and study the architecture of A1R allosteric site to target it at a molecular level. We then applied an algorithm to screen over 160 million compounds and narrowed the list to 26 top-ranked candidates for experimental testing, which ultimately led us to discover the new set of A1R selective PAMs.”
Co-first author Dr Nicolas Panel, from Uppsala University, said the success of the study was driven by developing a new approach to virtually screen drug candidates for challenging allosteric pockets.
“Unlike traditional drug-binding sites, the A1R’s allosteric site is shallow and faces the membrane, making it very difficult to target,” Panel said. “To overcome this, we used molecular dynamics simulations to model how the membrane environment affects pocket shape, then screened a large compound library against this structure. This allowed us to identify new chemical scaffolds that would have been missed by standard methods.”
Uppsala’s Professor Jens Carlsson, who co-led the computational strategy, added, “This work shows how structure-based drug discovery can now be extended to membrane-facing pockets that were previously considered undruggable.”
The next phase of the research will focus on further preclinical development of the lead compounds and testing of their efficacy in disease models of neuropathic pain and ischemia-reperfusion injury. The team hopes these findings will lay the groundwork for future clinical trials and the development of safer, more targeted therapeutics for conditions involving A1R signalling.

The Biochrom Ultrospec 30 Cell Density Meter, available from Pacific Laboratory Products, is a compact and portable handheld instrument with a rechargeable Li-ion battery for easy movement around the lab. Suitable applications include performing microbial growth phase analysis in bacterial and yeast cell suspensions; bacterial transformation; plasmid amplification; antibiotic studies; quality control; contaminant detection; precision fermentation; and cell growth assays.
The easy-to-use cell density meter enables the creation of standard curves, giving users the ability to generate and save cell- and condition-specific standard curves using absorbance values or McFarland standards to use for future quantification. Measurement data can be exported to a USB flash drive as a .csv file.

The device has a touchscreen display, with an intuitive menu arrangement that is easily navigated using the colour touchscreen. It measures just 170 x 210 x 50 mm and weighs 0.6 kg. Pacific Laboratory Products www.pacificlab.com.au
Proteomics company Evosep has launched Evosep Eno — a robust, highthroughput separation tool for nextgeneration workflow integration for LC-MS based proteomics. It should enhance Evosep’s ability to advance the use of LC-MS based proteomics as a vital tool for the pharmaceutical and biotech industries.

Designed to drive the capabilities of next-generation LC-MS based proteomics, the product offers good standardisation, high performance and ease of use. It should enable users to speed up drug discovery and translational research programs, while enhancing their capabilities to gain insights into the proteome quickly.
The platform can analyse more than 2000 proteins per minute at 500 samples per day, enabling throughput for large-scale studies. This should accelerate biomarker discovery and clinical research without compromising data quality.
It provides consistent and reproducible results across different systems and locations, meaning it offers trustworthy data for multi-site studies conducted by academic research centres, drug developers and contract research organisations.
The product’s standardised workflows with integrated consumable solutions are designed to simplify and streamline processes, which should increase efficiency and reduce errors.
Accurate Mass Scientific accuratems.com
The NL5+ confocal system is the second generation of fast line-scanning confocal technologies from Confocal.nl, providing high-contrast images from thicker specimens such as organoids, tissue samples, and plant and animal model organisms. It is designed to provide high temporal resolution, while at the same time reducing phototoxicity; since phototoxicity is one of the biggest challenges for researchers, this makes the device useful for live cell imaging.
The NL5+ module can be added to any widefield microscope to turn it into an advanced 3D fast-scanning confocal imaging system. The NL5+ system is available as a complete package with options for confocal, widefield or brightfield observations.
The combination of sCMOS camera-based detection with sensitivity up to 96% QE and a set of filters allows acquisition of images in up to six different colours; this should provide an improved temporal resolution, high sensitivity and good signal-to-noise ratio. Additional motorised single-band emission filters enable even faster imaging with cleaner results. Resolution in real time is 170 nm with deconvolution, while the FOV is 18 mm (FN18) with 330 x 330 µm (40x objective). The speed in line-scanning mode is up to 75 fps for 2048 x 512 px and up to 55 fps for 2048 x 2048 px.
Line-rescan microscopy using the system is suitable for biological applications where a combination of speed and high sensitivity is required. For example, it is possible to study organoids from different organs, such as the intestine, liver, kidney and brain. The product can provide information on morphology, cell differentiation, gene expression, signal transduction, and interactions of organoids with their environment. This can enable insights into the development, function and pathology of organs, and to new possibilities for personalised medicine. SciTech Pty Ltd www.scitech.com.au


Interchangeable parts service for singleuse assemblies
Watson-Marlow Fluid Technology Solutions (WMFTS) has introduced WMArchitect Interchangeable Parts to secure a resilient supply of single-use assemblies by including prequalified, alternative components in the design stage to avoid regulatory compliance interruptions and long lead times.
As per the service, the company works with its customers to assess form, fit and function for alternative components; interchangeable components are then advised during the early assembly design stage of the assembly specification. WMFTS proactively monitors the lead times of components to track whether there are gradual or sudden changes. When a component flags, the company can pivot quickly to source a pre-agreed alternative.
The bioprocessing service eliminates the disruption of reduced component stock and potential production shutdowns, to instead enable a simplified way to substitute in suitable components with shorter lead times. By agreeing on alternative component options during the design stage, customers can avoid the supply constraints of a specific component such as tubing, tri-clamps and other tube fittings. Biopharmaceutical companies are thus offered a flexible solution to the uncertainty of choosing substitute components and the absence of clear guidance in standardised criteria.
Interchangeable Parts is fully supported by experienced specialist scientists and a Validation Testing team. MFTS has adopted the BioPhorum best practice equivalency assessment method to address any concerns over compatibility, performance and safety. Full compliance and traceability is maintained, and by swapping in a component that is more easily available, disruption to workflow is limited.
WMArchitect single-use solutions is an end-to-end biopharmaceutical fluid management solution from a single supplier. It is designed to offer both standard, ready-to-use single-use assemblies as well as customised designs that are fully supported with tailored validation packages.
Watson-Marlow Fluid Technology Solutions www.wmfts.com/en-au/#

• LIQUID LEVEL indiCation
• INDIRECT leVel GauGeS
• BOILER WATER monitorinG
• GERmAN expertiSe & quality


The success of lipid nanoparticles (LNPs) in drug delivery — especially in the development of mRNA vaccines — relies on precise and reproducible characterisation. As the race to develop next-generation therapeutics intensifies, precision tools like the Malvern NanoSight Pro and Malvern Zetasizer Advance are proving pivotal. These technologies enable detailed measurement of critical parameters like particle size, polydispersity, concentration and zeta potential — all factors that can directly influence stability, consistency and efficacy of nanoparticle formulations.
While both systems analyse particle size, they do so using complementary methods that provide different insights. The Zetasizer is based on dynamic light scattering (DLS), which delivers rapid, intensity-weighted size measurements. Zetasizer Ultra enhances this with Multi-Angle Dynamic Light Scattering (MADLS), providing higher resolution of multimodal samples, enhanced repeatability and calibration-free particle concentration analysis. Its Adaptive Correlation technology uses statistical analysis to identify data points that are not representative of the main particle population, which should result in more trustworthy measurements particularly in the presence of aggregates.
In contrast, NanoSight Pro uses nanoparticle tracking analysis (NTA) to track individual particles in real time, yielding more accurate size distributions in polydisperse samples. Its intelligent software, AI-driven autofocus and video-based tracking make it faster, easier to use and more consistent across users. Beyond sizing, the product also measures particle concentration, enabling verification of LNP formation — an essential aspect of formulation development and quality control. With built-in fluorescence capability, NanoSight Pro allows selective tracking of labelled nanoparticles in complex biological media — suitable for exosome, virus and drug delivery research — to distinguish target particles from background noise, assess surface modifications, confirm payloads or evaluate the success of purification methods.
Together, these systems offer a novel cross-validation capability and high data confidence. By validating results across two orthogonal technologies, scientists reinforce particle size as a Critical Quality Attribute (CQA), influencing everything from in vitro performance to manufacturing outcomes.
ATA Scientific Pty Ltd www.atascientific.com.au

The TecSense HSA online O2 headspace analyser is a semiautomatic device that determines the O2, CO2 and foreign gas in the headspace of bottles. It is designed for use at the filling line for precise measurements.
A sample bottle or can is inserted into the HSA analyser and the measurements of O2 and CO2 are taken. Foreign gas is also detected when present in the bottle or can. The product further calculates the volume of the product within the bottle.
Key features include: determination of headspace in 30 s; determination of O2 content; determination of foreign gas; determination of CO2 content; calculation of the O2 concentration mg/L in the bottle; display of the amount of liquid in the bottle; digital data connection; a touch screen; and easy handling.
The product is useable at the TecServiceHSA filling station for direct usage on PC. It also offers the ability to save data on USB or directly via TecServiceHSA.
AMS Instrumentation & Calibration Pty Ltd www.ams-ic.com.au




Using a lab-grown material, researchers at the National University of Singapore (NUS) have revealed that some of the effects of aging in the heart may be slowed and even reversed. Their discovery, which has been published in the journal Nature Materials, could open the door to therapies that rejuvenate the heart by changing its cellular environment, rather than the heart cells themselves.
The team focused on the extracellular matrix (ECM) — the complex framework that surrounds and supports heart cells. This netlike scaffolding made of proteins and other components holds cells in place and sends chemical signals that guide how the cells function.
As the heart ages, the ECM becomes stiffer and its biochemical composition changes. These
changes can trigger harmful activity in heart cells, contributing to scarring, loss of flexibility and reduced function.
“Most aging research focuses on how cells change over time,” said team leader Assistant Professor Jennifer Young. “Our study looks instead at the ECM and how changes in this environment affect heart aging.”
According to PhD student Avery Rui Sun, it has until now been difficult to pinpoint whether physical stiffness or biochemical signals play the bigger role in
driving heart aging, because they usually happen at the same time. To investigate this, the team developed a hybrid biomaterial called DECIPHER (DECellularized In Situ Polyacrylamide HydrogelECM hybrid), made by combining natural heart tissue with a synthetic gel to closely mimic the stiffness and composition of the ECM.
“The DECIPHER platform … [allows] researchers to independently control the stiffness and the biochemical signals presented to the cells — something no previous system using native tissue has been able to do,” Sun said.
When the team placed aged heart cells onto DECIPHER scaffolds that mimicked ‘young’ ECM cues, they found that the cells began to behave more like young cells — even when the material remained stiff. Closer investigation revealed that this included a shift in gene activity across thousands of genes associated with aging and cell function. In contrast, young cells placed on ‘aged’ ECM began to show signs of dysfunction, even if the scaffold was soft.
“This showed us that the biochemical environment around aged heart cells matters more than stiffness, while young cells take in both cues,” Young said.
“Even when the tissue was very stiff, as it typically is in aged hearts, the presence of ‘young’ biochemical signals was enough to push aged cells back toward a healthier, more functional state. This suggests that if we can find a way to restore these signals in the aging heart, we might be able to reverse some of the damage and improve how the heart functions over time.
“On the other hand, for young heart cells, we found that higher stiffness can cause them to prematurely ‘age’, suggesting that if we target specific aspects of ECM aging, we might slow or prevent heart dysfunction over time.”
While the work is still in the research phase, the researchers said their findings open up a new direction for therapies aimed at preserving or restoring heart health during aging — by targeting the ECM itself. Beyond the heart, they believe the DECIPHER method could be applied to studying aging and disease in other organs as well, due to the major role of the ECM in cell function across all our tissues.
“Many age-related diseases involve changes in tissue stiffness — not just in the heart,” Young said. “For example, the same approach could be applied to kidney and skin tissue, and it could be adapted to study conditions like fibrosis or even cancer, where the mechanical environment plays a major role in how cells behave.”
Bio-Rad Laboratories has extended its range of recombinant monoclonal antiidiotypic antibodies with the introduction of antibodies specific to pertuzumab (Perjeta), guselkumab (Tremfya), canakinumab (Ilaris), belimumab (Benlysta) and the bispecific drug emicizumab (Hemlibra). The company has also extended its range of SpyCatcher reagents to include Human IgM-FcSpyCatcher.
Bio-Rad’s comprehensive range of anti-biotherapeutic antibodies enables the development of robust bioanalytical assays against biologic drugs and is suitable for use in pharmacokinetic (PK) and anti-drug-antibody (ADA) assays for pertuzumab, guselkumab, canakinumab, belimumab and emicizumab, facilitating therapeutic drug monitoring for innovator and biosimilar products.
Human IgM-FcSpyCatcher is a reagent that allows researchers to rapidly switch antibody formats at the protein level, enabling spontaneous formation of pentameric and hexameric IgMs. Compatible with the company’s recombinant HuCAL antibodies, the reagent has been successfully tested in ELISA, western blotting and precipitation assays.
The company’s antibodies and reagents are approved for in vitro research, with the anti-idiotypic antibodies also approved for commercial in vitro testing services to support preclinical and clinical drug and biosimilar development, and patient monitoring.
Bio-Rad Laboratories Pty Ltd www.bio-rad.com
Manual calibration and parameter input in systems are time-intensive and error-prone, posing significant challenges across industries. Studies reveal that manual data entry carries a 4% error rate, equating to as many as 400 errors in 10,000 entries. Additionally, employees can spend over 100 hours monthly on tasks like calibration parameters manual entry, consuming valuable resources.
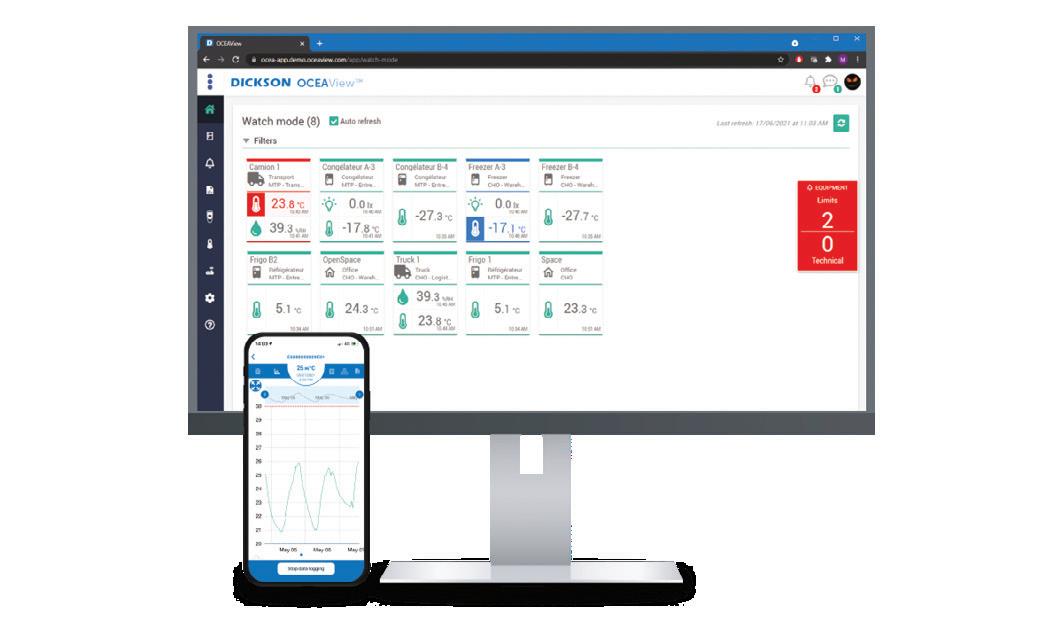
Ten years after the launch of its Replaceable Sensors, The Dickson Company is releasing Smart DS — its second generation of intelligent sensors, designed to deliver a smarter, easier to use and more autonomous monitoring solution. By embedding calibration and traceability data directly into each sensor, the device eliminates the need for full device recalibration.
The updated architecture allows for faster sensor replacement, uninterrupted monitoring and total compliance. Designed for critical environments like freezers, refrigerators, cleanrooms, laboratories and pharmaceutical storage, the smart sensors provide continuous tracking of ambient conditions, from temperature to humidity, differential pressure and CO2
This second generation of sensors is designed to enable seamless maintenance, reduce downtime and provide full traceability of calibration data. It includes digital temperature sensors; digital temperature and humidity sensors; 3-in-1 digital sensors for CO2, temperature and humidity; digital differential pressure sensors; 4–20 mA compatible smart sensors; and Smart PT100 temperature sensors.
Antidote Biomedical www.antidote.com.au
Mercodia’s Lp(a) ELISA provides a method for the quantitative determination of human lipoprotein(a) in serum or plasma. It is a solid phase two-site enzyme immunoassay based on the sandwich technique, in which two monoclonal antibodies are directed against separate antigenic determinants on the apolipoprotein(a) molecule. Apolipoprotein(a) in the sample reacts with anti-apolipoprotein(a) antibodies bound to microtitration wells and peroxidase-conjugated anti-apolipoprotein(a) antibodies in the solution.
Each ELISA kit contains reagents for 96 wells, allows for a sample volume of 25 µL, has an assay range of 0.3–5.0 U/L with a detection limit of 0.07 U/L, and has a total incubation time of 2 h and 15 min. Plasminogen and apolipoprotein B cross-reactions have not been detected.
Mercodia’s Human Lp(a) ELISA kits are distributed in Australia and New Zealand by Sapphire Bioscience. Sapphire Bioscience www.sapphirebioscience.com
Almost 90% of people who use pipettes for more than an hour a day report discomfort — from the thumb, wrist or arm, all the way up to the shoulder and neck1 — due to prolonged and repetitive movements. Pipette design has a significant influence over operator body position and force required while pipetting, as well as how often a movement needs to be repeated. For instance, lightweight, wellbalanced pipettes are easier to hold, while finger hooks ensure that some of the pipette’s weight rests on the index finger, allowing a looser, more relaxed grip.
When using mechanical pipettes, the plunger needs to be manually depressed and released to aspirate and dispense liquids. Traditional pipettes also use a rotating plunger or dial to set pipetting volumes, making frequent volume adjustment tiring for the wrist and fingers.
INTEGRA Biosciences’ EVOLVE manual pipettes feature 3 separate quick set dials — enabling users to set volumes more than 10 times faster — and have minimal plunger forces and stroke distances to minimize the risk of repetitive strain injuries (RSIs) such as thumb tenosynovitis.
Using multichannel pipettes can help to reduce the number of repetitive movements required for tasks like transferring reagents from a reservoir to a microplate. For example, instead of performing 96 transfers from the reservoir to the 96 well plate with a single channel pipette, an 8 channel pipette will reduce the number of transfers to just 12. The EVOLVE is available in single, 8, 12 and 16 channel options to minimize
the number of pipetting steps, with a rotating handle on all multichannel options.
fatigue with electronic solutions
Electronic pipettes reduce hand and thumb strain compared to mechanical pipettes, as liquid displacement is controlled by a motor. They also offer repeat dispensing functions, which reduce the time spent pipetting by allowing the user to aspirate a larger volume of liquid, then aliquot it into several separate wells or vials. INTEGRA’s VIAFLO electronic pipettes provide 10 intuitive, preset pipetting programs — including repeat dispensing — to accelerate and simplify liquid handling workflows. Pipetting parameters can be easily adjusted using an ergonomic thumbwheel interface, eliminating the need to repeatedly push buttons to change settings, further lowering the likelihood of strain or injury.
Adjustable tip spacing multichannel pipettes are ideal for transferring samples between different labware formats in parallel, such as between tubes and 384 well plates. VOYAGER electronic multichannel pipettes offer a unique automatic adjustable tip spacing function, allowing you to expand or reduce tip spacing at the push of a button. This enables effortless labware reformatting and drastically cuts the time spent pipetting, also reducing the risk of RSIs.
minimizing
INTEGRA’s 96 and 384 channel benchtop pipettes eliminate the need to manually lift and stabilize pipettes altogether, using servo
motor-assisted movements to make pipetting effortless. These pipettes can fill entire 96 or 384 well plates in one step, greatly increasing efficiency. The ASSIST PLUS pipetting robot goes one step further, eliminating physical strain by fully automating repetitive liquid handling tasks. This affordable solution can automate VIAFLO and VOYAGER electronic pipettes, as well as D-ONE single channel pipetting modules, and is simple to program with VIALAB software for maximum workflow flexibility.
Ergonomic pipettes enhance productivity
It’s clear that laboratories can improve efficiency while significantly reducing the risk of RSIs by opting for ergonomically designed liquid handling tools. Investing in the right pipetting solutions — from user-centered manual pipettes to fully automated benchtop systems — doesn’t just increase productivity, it protects the long-term well-being of lab professionals. Ready to experience pain-free pipetting? Then book a free online or onsite training session with one of INTEGRA’s liquid handling experts today! You can also request a product demo to learn more about ergonomic pipetting and INTEGRA’s solutions.
1. Analysis of the musculoskeletal loading of the thumb during pipetting – A pilot study - PMC
Faruhi,
In 2025, academia is facing an incredible amount of political and financial pressure. Following US President Donald Trump’s return to the Oval Office in January, universities face further budget cuts against the backdrop of a lack of trust from reliable donors and scrutiny over ideological bias. As respected institutions continue to face cuts to their funding, there is a question mark over both the volume and quality of research that can be conducted — and research integrity may be essential to academia’s very survival.
Due to the new federal funding policies, including cuts to National Institute of Health (NIH) overheads, suspended grants and increased political scrutiny, academic institutions now face a very real financial shift. Since Trump’s return to office, university funding has been in the spotlight as part of a wider focus on perceived waste, illustrated by the establishment of the Department of Government Efficiency (DOGE). The administration has suspended federal grants to Princeton University from the Department of Energy, NASA and the Department of Defense, while Harvard University has seen up to $9 billion in federal funding come under the spotlight.
In addition, the NIH said it would be cutting billions in overhead payments by capping so-called indirect costs for researchers at 15%, which has been described as “a sure-fire way to cripple lifesaving research and innovation”1. Overheads typically cover essential research costs, including equipment, administration and researcher salaries. At the same time, negative media coverage of campus
controversies has further affected institutions. Additionally, accusations have emerged that the Trump administration’s funding cuts are partly motivated by perceptions that conservative values are underrepresented in academia.2
The NIH’s decision to cap overhead support at 15% sparked strong backlash from leading research institutions, prompting a federal lawsuit aimed at reversing the rule change. This legal action underscores the scientific community’s profound concern over the funding cuts, adding another layer of uncertainty for academic institutions already struggling to maintain essential research operations.3
A vicious cycle
As a result of these funding cuts, research integrity has become even more central — if it wasn’t already — to institutional credibility and financial stability. Research Integrity Officers (RIOs) and journal editors have warned that errors, even those that aren’t malicious but could lead to paper retractions, can seriously damage an institution’s reputation and funding. With federal support shrinking, donations have become the major funding source, making it essential to

maintain the trust of donors, who care deeply about institutional credibility. Any integrity issues could risk reputational harm that deters donations and threatens long-term survival.
Research integrity, therefore, is no longer a compliance obligation, but an existential necessity to the survival of these academic institutions.
Current funding policies from the Trump administration mean that covering basic research costs, including equipment, administration and salaries, is increasingly difficult, putting more stress on universities to plead for more support from donations. Nobel Prize winners are among 2000 American scientists who warned that independent research is being jeopardised because of cuts and a “climate of fear”.4
The renewed funding pressures come not long after the NIH has undergone reforms to promote transparency in the research process, including with its Data Management and Sharing Policy, which requires grant applicants to publicly share their data in a timely and transparent manner, as well as guard against any plagiarism, image or statistic misuse.
To safeguard research integrity, we recommend using a suite of quality-assurance

tools. Proofig AI, for example, automatically screens images for manipulation, duplication, image plagiarism and AI-generated images. Another widely used tool is iThenticate, which helps detect text plagiarism before submission. In addition, electronic lab notebooks (ELNs) like LabArchives or Biodata support good practices by organising data and maintaining version history. Together, these quality-assurance tools help uphold standards across research outputs and guard against errors that can lead to retractions or reputational damage. While these solutions help uphold integrity, the broader funding pressures are already reshaping the global research landscape.
Funding pressures in the United States have prompted top American scientists to seek opportunities abroad, particularly in fields seen as ideologically sensitive — such as climate change, diversity and vaccines. According to the Financial Times5, institutions in Europe and China are actively courting US-based researchers. The president of the EU’s European Research Council has called the US climate “discouraging for independent investigator-driven research”,
reflecting broader concerns that academic freedom and integrity are increasingly at risk.
The increasingly ideologically driven climate means researchers face tighter scrutiny and stricter rules, making effective tools to safeguard research integrity more crucial than ever. The consequences of failing to safeguard research integrity are no longer theoretical — they are playing out in real time, with serious financial and reputational fallout.
In recent years, high-profile cases at leading universities and research institutions — including a $112.5 million settlement for research misconduct6, fraudulent HIV vaccine funding scandals totalling $7.2 million 7, and controversies involving prominent figures 8 at top institutions9 — have demonstrated the staggering financial and reputational costs of compromised research integrity10. In a political climate where funding for research is being increasingly eroded, ensuring research integrity is no longer optional — it is a survival imperative for institutions seeking to protect their reputation and secure critical funding.
Tools like Proofig AI for image integrity and iThenticate for text plagiarism offer essential
safeguards against these catastrophic risks by providing automated, reliable integrity checks across different aspects of research output. These complementary solutions help institutions maintain trust, avoid devastating financial losses, and uphold the highest standards of scientific excellence. As funding challenges intensify in 2025, protecting research integrity has never been more critical.
1. https://www.science.org/content/article/scienceadviserslashing-nih-overhead-payments-sparks-outrage<
2. https://www.nytimes.com/2025/04/14/us/politics/ trump-pressure-universities.html
3. https://www.science.org/content/article/can-nihoverturn-court-order-blocking-it-slashing-overheadpayments-unlikely-one-expert
4. https://www.npr.org/2025/04/01/nx-s1-5347411/ scientists-trump-research-national-academiesopen-letter
5. https://www.ft.com/content/cdcbe3df-9475-48169a95-0df64838566f
6. https://www.justice.gov/archives/opa/pr/dukeuniversity-agrees-pay-us-1125-million-settle-falseclaims-act-allegations-related
7. https://apnews.com/16f04abe33e647228cb50d567 09c93d7
8. https://news.mit.edu/2005/misconduct
9. https://www.thecrimson.com/article/2024/3/15/ginoharvard-investigation-report/
10. https://www.nature.com/articles/d41586-022-02002-5

The Haenyeo, a group of all-female divers from the Korean island of Jeju, are renowned for their ability to dive in frigid waters without the aid of breathing equipment — even while pregnant. An international research team has now found that the divers’ remarkable abilities are due to both training and genetic adaptation, with their results published in the journal Cell Reports
The Haenyeo, or ‘women of the sea’, dive year-round in social collectives to harvest food for their communities. They begin training at around age 10 and continue for their whole lives. Inspired by the Haenyeo’s remarkable diving abilities, the researchers wanted to know whether they have distinguishable physiological traits that help them cope with the strain of diving, and if so, whether these traits are due to genetic adaptation or training.
To find out, the team compared the physiological traits and genomes of 30 Haenyeo divers to 30 non-Haenyeo people from Jeju, as well as 31 people from mainland Korea. To match the age of the divers, the average age of all participants was 65. The researchers compared the participants’ heart rate and blood pressure at rest and during ‘simulated dives’ where the participants held their breath while submerging their faces in cold water.
“If you hold your breath and put your face in a bowl full of cold water, your body responds as if you’re diving,” said lead author Melissa Ilardo, from The University of Utah. “A lot of
the same processes happen in your body that would happen if you were to jump in the ocean, but it’s done in a way that’s safe for people with no diving experience.”
The team’s genomic analysis showed that Jeju residents — both Haenyeo and non-Haenyeo — were distinct from individuals from mainland Korea, suggesting that all Jeju residents are descended from the same ancestral population. Ilardo explained, “We can essentially think of everyone from Jeju as either ‘diving Haenyeo’ or ‘non-diving Haenyeo,’ because their genetics are the same.”
The genomic analysis also revealed two gene variants in the Haenyeo that may help them cope with the pressures of diving, making the Haenyeo the second known population of traditional breath-hold divers that has evolved for diving. One gene is associated with cold tolerance, which could make the divers less vulnerable to hypothermia. The other gene is associated with decreased diastolic blood pressure (ie, blood pressure in between heart contractions). The variant was found in 33% of participants from Jeju but only 7% of mainland participants.
“This association may reflect natural selection to mitigate the complications of diastolic hypertension experienced by female divers while
diving through pregnancy,” Ilardo said. “Since Bajau women also dive while they’re pregnant, we wonder whether pregnancy is actually driving a lot of the genetic changes in these diving populations.”
During the simulated dives, all of the participants showed decreased heart rates, but the Haenyeo’s heart rates dropped significantly more than those of either control group. On average, the divers’ heart rates decreased by 18.8 bpm compared to a decrease of 12.6 bpm in the Jeju non-divers. A lowered heart rate during diving is beneficial because it saves energy and conserves oxygen. Since their genomic analysis indicated that Haenyeo and non-diving Jeju are genetically members of the same population, the researchers concluded that this feature is likely due to the divers’ training.
“Because the Haenyeo have been diving for a very long time, their heart rate has been trained to drop more,” Ilardo said. “This was something we could actually visually see — we had one diver whose heart rate dropped by over 40 beats per minute in less than 15 seconds.”
The researchers said their findings highlight the potential of studying traditional diving populations to better understand human genetic and physiological adaptation.
“We’re really excited to learn more about how these genetic changes may be affecting the health of the broader population of Jeju,” Ilardo concluded. “If we can more deeply characterise how those changes affect physiology, it could inspire the development of therapeutics to treat different conditions, such as hypertensive disorders of pregnancy and stroke.”
Peter Davis, ATA Scientific
It is not every day that you get to solve a unique problem. But that’s exactly what happened when a collaboration with researchers at the University of Melbourne led to a world first breakthrough in scalable Metal Phenolic Network (MPN) nanoparticle production.
About a year ago at the 14th Nanomedicines Conference on Sydney Harbour, I was one of the fortunate people to witness the presentation by Prof Frank Caruso of Chemical Engineering, Melbourne University, describing the effectiveness of Metal Phenolic Networks (MPNs) to target specific organs. This science is extraordinary given it has the potential to create organ specific nanoparticles for drug delivery.
However, translating this science into scalable production posed significant hurdles. The method can be very labour intensive, and particularly difficult to automate as there are multiple order and time sensitive components required.
During discussions with postdoctoral researchers Dr Christina Cortez-Jugo and Dr Qingqing Fan, it became clear that existing off the shelf technologies that have been developed for LNPs couldn’t easily handle the unique requirements of newer MPN formulations. Specifically, the requirement to sequentially introduce multiple distinct components with precisely controlled timing and order presented a challenge not addressed by current commercial technologies.
Micropore Technologies have already solved many issues of scaling complex chemistries in the past. ATA Scientific created a test rig with the Pathfinder system and some very cool modifications at our disposal. Theories around flow dynamics and introduction speed were numerous, and at times perplexing. In practice, with the help of Dr Jingqu (Rachel) Chen we simply set up our best estimate to see if we could even make the nanoparticles. Post processing and subsequent analysis on the Malvern Panalytical Zetasizer Ultra Red proved we had extraordinary success!

An experiment plan followed, to demonstrate the scalability of MPN NPs using Pathfinder. The following was proposed:
1. Batch comparison: Produce both manual and Pathfinder-fabricated MPN NPs using 2–3 well-reported formulations with varying components. These will be scaled at 2 mL, 5 mL, 10 mL, 50 mL, and 100 mL volumes, with comparison of size, zeta potential, encapsulation efficiency, and in vivo performance across batches.
2. Functional testing: Preparation of largerscale batches of MPN nanoparticles loaded with various therapeutic small molecule drugs using the Pathfinder system to evaluate functional performance. Results will be benchmarked against manually synthesized nanoparticles using identical analytical parameters.
Why does this matter?
Without scalable and reproducible production, the entire MPN project could have remained stuck in the lab. It is now clear that the flexibility of the Micropore Technologies Pathfinder uniquely resolves this challenge — not just at lab scale, but full GMP production is clearly possible given this technology is used to create
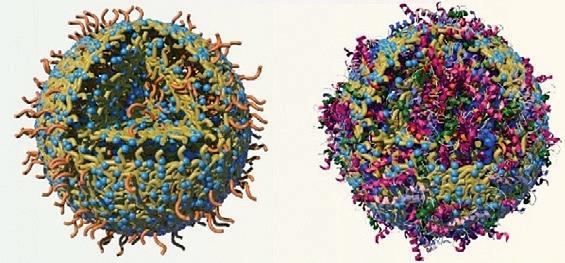
Cargo-loaded MPNs NPs. Image has been cropped from the original (doi.org/10.1002/anie.202312925) and is courtesy of the study authors under CC BY 4.0
litres/minute, opening the opportunities for treatments with the MPN technology.
What’s next?
Buoyed by the preliminary results, the team have submitted an abstract to the A-RNA25 conference later this year entitled “Bioactive metal-organic nanoparticles: a versatile platform for RNA delivery”. This is a presentation not to be missed — visit www.a-rnameeting. org.au for more details.
For more information on how the Pathfinder may solve your nanoparticle needs, contact Pete Davis, ATA Scientific +61 0417 778 971 or email pdavis@atascientific.com.au
ATA Scientific Pty Ltd www.atascientific.com.au

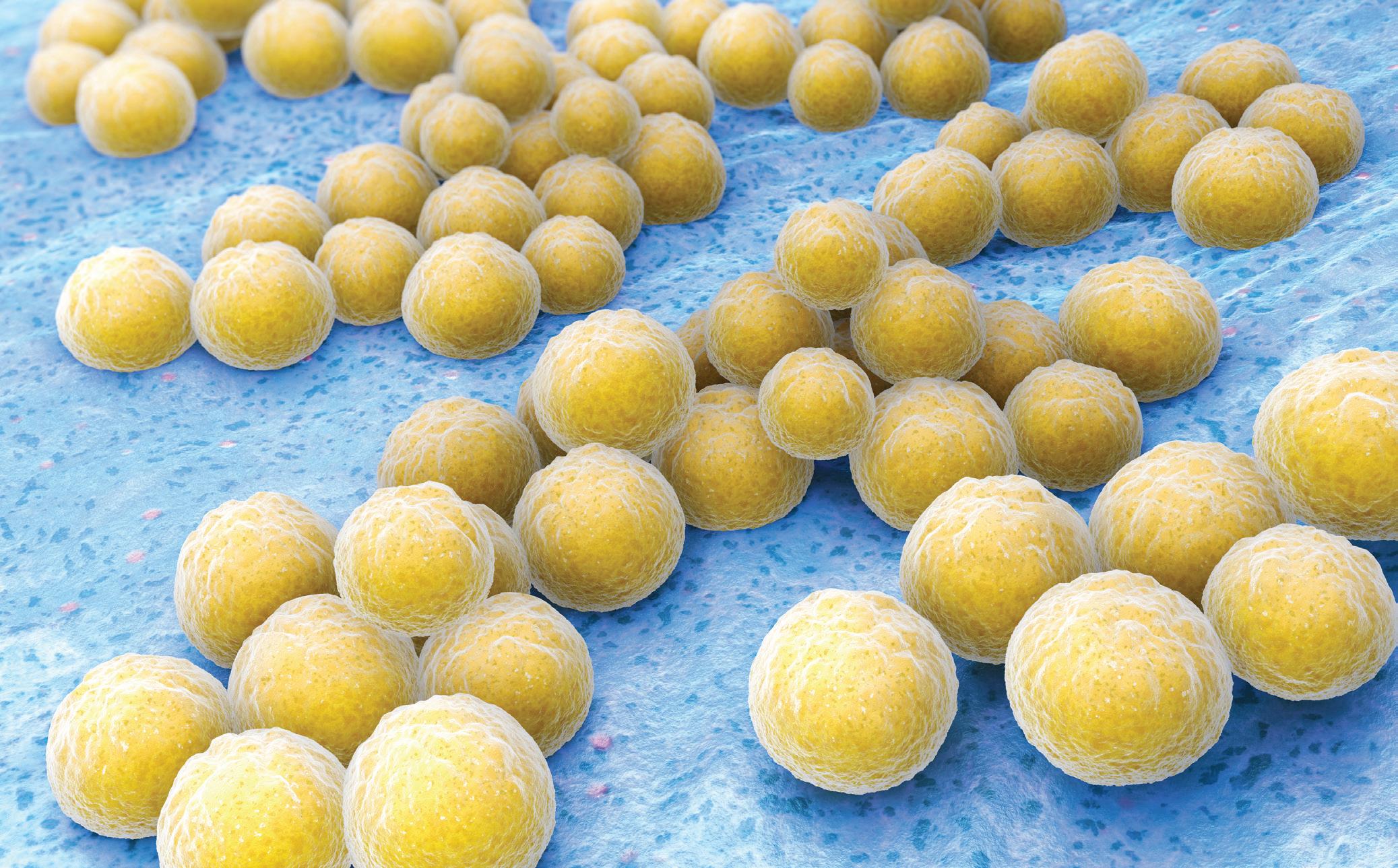
A research team led by The Peter Doherty Institute for Infection and Immunity has taken a major step towards using real-time genome sequencing in routine hospital care, showing that tracking bacterial changes during serious Staphylococcus aureus (golden staph) infection can give clinicians immediate insights to personalise treatment and improve patient outcomes.
Golden staph is a superbug that is responsible for more than one million deaths globally each year. These bacteria can cause lifethreatening infections such as sepsis, pneumonia, bone and joint infections, and endocarditis (infection of the heart valves), and are known for their ability to adapt quickly — becoming resistant to even the most potent antibiotics.
Unlike standard hospital laboratory tests that only identify the type of bacteria, genome sequencing reveals a complete genetic profile — including traits that influence its response to treatment. But studies on bacterial evolution during infection have until now only been conducted retrospectively, often years after treatment.
The new research in Nature Communications saw scientists from the Doherty Institute, in partnership with seven major Victorian hospitals, collect golden staph samples from patients with severe, recurring infections at the time of treatment failure. These strains were compared to strains obtained at the beginning of the infection, with the findings returned to the treating physicians. In one-third of the cases, the bacteria had picked up dangerous mutations that made treatment more likely to fail.
“In one case, after initially controlling a golden staph infection, the patient returned to hospital two months after stopping antibiotics,” said study leader Dr Stefano Giulieri, an infectious diseases physician and clinician researcher at the Doherty Institute.
“Samples were referred to us for sequencing and we discovered that, during infection, the golden staph bacteria had become 80 times more resistant to the antibiotic used. Each time they reappeared in blood, the bacteria had picked up a new dangerous mutation.
“Using this information, the clinical team was able to choose a new treatment that was able to finally cure the infection.
“Our study is the first to show that by tracking bacterial evolution in real time, genome sequencing can reveal tricks bacteria use to survive, giving doctors the power to stay one step ahead and tailor treatment to the specific bacterial strain. This helps avoid unnecessary treatments,
minimise side effects for patients and prevent further antibiotic resistance — ultimately giving patients the best chance of recovery.”
To assess the value of this approach, the researchers surveyed 25 infectious disease specialists across Australia, Switzerland, the UK and the US. Clinicians rated the genomic reports as ‘highly useful’ (80 out of 100) and indicated that the information influenced their choice of antibiotic treatment in more than a third of cases — with survey participant Professor Eugene Athan, an infectious diseases consultant at University Hospital Geelong, saying the approach offers the opportunity for infectious diseases to enter the era of precision medicine, just as cancer genomics has done in oncology.
“The ability to track bacterial evolution in real time during severe infections is a game changer for clinicians,” Athan said.
Professor Benjamin Howden, Director of the Microbiological Diagnostic Unit Public Health Laboratory (MDU PHL) at the Doherty Institute and senior author of the study, said the findings “represent a major step toward targeted therapy for bacterial infections and open the door to future clinical trials that could make this approach standard practice in hospitals worldwide”. The MDU PHL is now exploring a new service to provide advanced genomic investigations for cases where treatment is failing, which would result in Victorian hospitals becoming the first in the world to access precision microbiology in clinical care.
Lonza’s 4D-Nucleofector LV Unit PRO is a next-generation, large-scale electroporation unit designed to deliver clinically relevant cargos into large volumes of T cells. The unit builds on the company’s non-viral large-scale transfection platform and is optimised for CRISPR-based genome engineering and other advanced cell therapy applications.
For more than 20 years, Lonza’s Nucleofector Technology has been used in cell and gene therapy R&D, enabling efficient delivery into hard-to-transfect primary cells, such as T cells. The 4D-Nucleofector LV Unit PRO further advances this technology, offering enhanced performance for CRISPR-based knock-out, knock-in and transposition.
A key innovation is the implementation of the Nucleocuvette Cartridges PRO, an improved cartridge version that enables the electroporation of complex cargos into up to one billion cells per run. The updated cartridge supports complex clinically relevant cargo delivery and introduces several usability improvements, including greater flexibility, simplified loading and clearing, and increased robustness of the flow-through process.

The upgraded system is suitable for preclinical studies and process development, serving as a scalable and reproducible platform for a seamless translation from discovery projects to GMP-compliant manufacturing for next-generation cell and gene therapies.
Scientifix Pty Ltd
www.scientifix.com.au

TOPTICA’s DL pro BFY tuneable diode laser is especially suitable for applications where carefree laser operation is required. The hermetically sealed butterfly package and the compact design of the resonator allow for the best passive mode stability and robustness within the DL pro family. It can even run as an outdoor system, equipped with temperature stabilisation, to withstand massive winds and pressure fluctuations.
The DL pro range of ECDL lasers is suitable for almost all wavelengths between 369 and 1770 nm. With power up to 400 mW and free running linewidth down to 0.6 kHz, the laser is suitable for most QT applications. Its digital controller allows a stable performance, the GUI making remote access easy, which is convenient when the DL pro rack version is requested. It is fully compatible with the extensive TOPTICA portfolio of tuneable diode lasers, driving electronics, software and accessories. Lastek Pty Ltd www.lastek.com.au
New England Biolabs (NEB) has launched the NEBNext Low-bias Small RNA Library Prep Kit to minimise biased representation of small RNA species in sequencing data. The kit’s next-generation small RNA preparation method is designed to be faster and less biased, with a broader input range than other commercially available kits.
Researchers have only recently realised that small RNA species — including rare and non-coding RNAs — have an outsized influence on health and disease. However, sequencing bias in pre-existing library preparation methods has made it hard for researchers to understand the true diversity and abundance of small RNAs in their samples. The prep kit was designed to generate RNA libraries with a more accurate representation of small RNA species.
Protocol enhancements include the addition of a novel splint adaptor that increases the diversity of interactions, facilitating ligation and increasing sensitivity, with a streamlined, simplified protocol. As a result, researchers can now analyse all RNA species present in biologically relevant samples. Additional improvements include high speed (~3.5 h), long shelf life (18 months) and wide input range. Standard and 2´-O-methylated samples can also be processed using the same protocol, with multiplexing enabled through up to 480 compatible UDI primer pairs (available separately).
The kit’s novel approach to adaptor ligation has culminated from over a decade of research by NEB scientists. The first study, published in 2012, recognised that RNA sequencing bias was significant and caused mostly by unpredictable differences in ligation efficiency for small RNAs. The second foundational study, published in 2020, presented a novel library preparation workflow that reduced bias and increased sensitivity in small RNA library prep. Several protocol enhancements have since improved the kit to outperform the originally published methods. New England Biolabs www.neb.com
Researchers at the US National Institute of Standards and Technology (NIST) claim to have developed a new and faster method for detecting and measuring the radioactivity of minuscule amounts of radioactive material.
Their innovative technique, known as cryogenic decay energy spectrometry (DES), could have far-reaching impacts, from improving cancer treatments to ensuring the safety of nuclear waste clean-up. It has been described in the journal Metrologia
Rapid detection and measurement
Traditional methods have proved effective at either measuring the amount of radioactivity or identifying which radioactive atoms are present — not both. Fully characterising a sample thus required the use of multiple techniques and intricate procedures using additional materials called tracers or calibrants.

The new method offers a streamlined approach, identifying radioactive elements and quantifying their level of radioactivity in even tiny samples — without needing extra materials. This allows scientists to better monitor, use and safeguard radioactive materials that affect public health and safety.
“Instead of waiting months for results, we can now get a full radioactivity profile in just a few days from a tiny sample,” said NIST physicist Ryan Fitzgerald.
The key to the technique is a transition-edge sensor (TES), a high-tech device widely used to measure radiation signatures. The TES provides
the capability to record individual radioactive decay events, in which an unstable atom releases one or more particles. By building up data from many individual decays, researchers can then identify which unstable atoms, known as radionuclides, produce the events.
“The TES is much more advanced than a familiar Geiger counter or other detectors used today,” Fitzgerald said. “Instead of just clicking to indicate radiation, or giving a blurry indication of the decay energy, it gives us a detailed fingerprint of what’s there.”
The TES device operates at extremely low temperatures, near absolute zero. When a radioactive decay occurs in a sample, the energy released is absorbed by the TES. This absorbed energy causes a tiny change in the electrical resistance of the TES.
The researchers precisely measured this change in resistance, which provides a highresolution ‘energy signature’ of the decay event. By analysing the detailed energy spectrum from multiple events, the researchers can identify the specific radioactive atom undergoing decay. This is possible because different radioactive atoms release unique energy signatures when they decay.
Inkjet precision
In their method, the researchers use a specialised inkjet device to carefully dispense tiny amounts, less than 1 millionth of a gram, of a radioactive solution onto thin gold foils. These gold foils have a surface dotted with tiny pores just billionths of a metre in size. These nanopores help to absorb the tiny droplets of the radioactive solution.
By precisely measuring the mass of the solution that was dispensed using the inkjet and then measuring the radioactivity of the dried sample on the gold foils, the researchers can calculate the radioactivity per unit mass, or the ‘massic activity’, of the sample. This inkjet method allows them to work with extremely small amounts

of radioactive material and still get an accurate measurement of its radioactivity.
The potential applications are vast. In medicine, this technology could help ensure the purity and potency of radioactive drugs used in cancer treatments. For nuclear energy, it could quickly identify radioactive composition of reprocessed fuel, speeding development of new advanced reactors.
Redefining radioactivity measurement
The newly reported research is the first step in a larger effort, known as the True Becquerel

Close-up of a superconducting sensor board containing multiple transition edge sensors (top row of squares), which detect energy released by individual radioactive decay events.
(TrueBq) project, to transform how we monitor and characterise radioactivity. The project aims to develop a more comprehensive measurement system that can handle a wide range of radioactive substances, including complex mixtures. It will integrate a precision mass balance system with the TES device to measure the massic activity of radioactive materials with unprecedented accuracy.
This new approach is said to represent a significant improvement over traditional workflows, which often involve multiple methods, chemical processing, and the use of chemical
tracers and standards. By streamlining the measurement process, TrueBq is expected to reduce the time required for analysis while simultaneously increasing accuracy.
While TrueBq’s current focus is on improving measurements at NIST itself, researchers have long-term aspirations for the technology. In the future, they hope to develop more portable and user-friendly versions of the system that could be deployed outside of NIST for critical applications in fields such as medicine, environmental clean-up and nuclear waste management.


Two separate research groups have been investigating the potential of tiny antibodies, sourced from the family of land animals known as camelids, to protect against certain viruses. Camelids include species such as camels, llamas and alpacas, and produce singledomain antibodies (also known as VHHs or nanobodies) that are much smaller than the antibodies generated by most animals, including humans.
One of the teams, led by Professor Daniel Watterson and Dr Ariel Isaacs from The University of Queensland (UQ), claims to have identified the first ever nanobody to work against Hendra and Nipah viruses — two highly lethal henipaviruses which have jumped from animals to humans in Asia and Australia. Their work has been described in the journal Nature Structural & Molecular Biology
First identified in Brisbane in 1994, Hendra virus has infected people via horses and flying foxes in eastern Australia, and was back in the headlines recently when a horse in South East Queensland died after contracting the state’s first case of the disease since 2022. Nipah virus outbreaks in people occur almost annually in Bangladesh and occasionally in other Asian countries where it is carried by bats. There are no approved human vaccines for either disease, nor are there any known cures — but with the
discovery of the new nanobody, that could all be about to change.
“A nanobody is one-tenth the size of an antibody, and being that small it can access hard-to-reach areas of a virus to block infection,” Isaacs explained.
“Nanobodies are also easier to produce and more stable at higher temperatures than traditional antibodies, so we are very excited about the potential of our discovery to lead to new treatments.”
The nanobody, called DS90, was among a series isolated by research partners at Universidad Austral de Chile from the immune cells of an alpaca called Pedro. DS90 was identified via a platform developed by Professor Alejandro Rojas-Fernandez which can isolate nanobodies against viruses of concern, with the researcher saying “we aimed to construct a broad barrier against future pandemic viruses based on scalable antiviral nanobodies — this fantastic work is just the beginning”.
Tests at Watterson’s laboratory at UQ’s School of Chemistry and Molecular Biosciences

confirmed DS90 could bind successfully to proteins in Hendra and Nipah viruses and block their ability to enter cells. The team used cryogenic electron microscopy at UQ’s Centre for Microscopy and Microanalysis to examine the process.
“We could see exactly how the nanobody bound to the virus reaching right into deep pockets, whereas antibodies typically just bind to exposed surfaces of viruses,” Watterson said.
“This new information is a crucial step towards using a nanobody to combat Hendra and Nipah, which cause outbreaks in people and can often lead to fatal respiratory and neurological disease.”
The team also combined the DS90 nanobody with a developmental antibody therapy that is

used as a last resort treatment for people infected with Hendra and Nipah.
“Excitingly, we demonstrated that the combination of DS90 with the m102.4 antibody — which is made at UQ — prevents Nipah virus from mutating and evolving,” Isaacs said.
“This is a powerful technique to prevent new deadly variants emerging.
“Other nanobodies have been approved for use as cancer treatments and it is now exciting to see that nanobodies can also be used to neutralise viruses.
“The next step will be to translate our findings into a therapeutic to be clinically ready in case of an outbreak of Hendra in Australia or Nipah in Asia.”
The second research team, based in Belgium, has meanwhile discovered a novel class of

nanobodies that are strongly protective against a wide range of SARS coronaviruses — including SARS-CoV-1 and numerous early and recent SARS-CoV-2 variants — offering a promising route to developing broad-spectrum antiviral treatments that could remain effective against future viral variants. Their findings have been published in the journal Nature Communications SARS-CoV-2, the virus behind COVID-19, continues to be a potential threat as it evolves into newer variants that are resistant to currently approved antibody therapies. Resistance largely emerges because antibodies typically target virus regions, such as the receptor binding domain of the spike protein, that also frequently mutate, enabling escape from antibody recognition.
To address this, a team led by Professor Xavier Saelens and Dr Bert Schepens at the VIB-UGent Center for Medical Biotechnology explored a different strategy by focusing on one of the more stable subunits of the spike protein. The so-called S2 subunit is critical for the virus’s ability to fuse with host cells, a process essential for infection, and it is more conserved across different coronaviruses. The team worked together with researchers from the VIB-VUB Center for Structural Biology, the VIB-UGent Center for Inflammation Research and VIB NANOBODY VHH Core (under the umbrella of VIB Technologies).
The researchers identified several antibodies that strongly neutralise a broad panel of SARS coronaviruses in a llama named Winter, who had been injected with stabilised spike proteins
from SARS-CoV-1 and MERS-CoV viruses back in 2016. What makes the antibodies in question particularly promising is their mode of action: they act like a molecular clamp. They latch onto the poorly exposed, highly conserved region (a coiled coil of three alpha helices) at the base of the virus’s spike protein. In doing so, they lock the spike protein in its original shape, physically preventing it from unfolding into the form the virus needs to infect cells.
The antibodies showed strong protection against infection in lab animals, even at low doses. When the researchers attempted to force the virus to evolve resistance, the virus struggled, producing only rare escape variants that were much less infectious. This points to a powerful, hard-to-evade treatment option.
“This region is so crucial to the virus that it can’t easily mutate without weakening the virus itself,” said Schepens, who was senior author on the study. “That gives us a rare advantage: a target that’s both essential and stable across variants.”
The team’s discovery thus marks an advancement in the quest for durable and broadly effective antiviral therapies, offering hope for treatments that can keep pace with viral evolution.
“The combination of high potency, broad activity against numerous viral variants, and a high barrier to resistance is incredibly promising,” Saelens said. “This work provides a strong foundation for developing next-generation antibodies that could be vital in combating not only current but also future coronavirus threats.”
76th
September 29–October 3, Sydney
The 76th International Astronautical Congress will see the world’s space community gather to access the latest advancements and trends, academic works, industry connections, and partnership opportunities.
IAC 2025 presents a generational opportunity to showcase the rapid progress of Australia’s space industry and the importance of space-enabled services to the Australian way of life. The event will delve into why space is critical for both businesses and our everyday lives, with the aim of advancing space in the Indo-Pacific by showcasing the entire region and its wide variety of space-based opportunities.
‘Sustainable Space: Resilient Earth’, the guiding theme of IAC 2025, explores the essential connection between space innovation and sustainability, focusing on three central areas: spacebased applications for Earth; sustainable space activities; and sustaining life beyond Earth. www.iac2025.org
Falling Walls Lab Australia Finale September 1, Canberra www.science.org.au/news-and-events/events/ falling-walls-lab-australia-2025
A-RNA 2025
September 1–3, Sydney www.a-rnameeting.org.au
Science at the Shine Dome 2025 September 1–4, Canberra aas.eventsair.com/science-at-the-shine-dome-2025
ASCIA 2025 Conference September 2–5, Brisbane and online ascia2025.com
BioProcessing Network 2025 Conference September 15–17, Melbourne www.bioprocessingnetwork.org.au/conference
IVC-23: 23rd International Vacuum Congress September 15–19, Sydney vacuumau.clubexpress.com/content.aspx?page_ id=22&club_id=875610&module_id=726931
ASC’s 52nd Annual Scientific Meeting September 26–28, Perth www.cytology.com.au/Web/Web/Events/AnnualScientific-Meeting.aspx
ASBMB 2025
September 29–October 1, Brisbane www.asbmb2025.com.au
Earth Science Week 2025 October 12–18, Australia-wide www.ga.gov.au/about/earth-science-week
AACB 62nd Annual Scientific Conference
October 13–16, Auckland aacb.eventsair.com/aacb-62nd-annual-scientificconference
Australasian Radiation Protection Society (ARPS) 2025 Conference October 19–22, Melbourne arpsconference.com.au
AusBiotechInvest 2025 October 21, Melbourne www.ausbiotechinvestment.com.au/flagshipevents-ausbiotechinvest
AusBiotech International Conference 2025 October 21–24, Melbourne www.ausbiotechnc.org
Blood 2025 and ISBT Perth 2025 October 26–29, Perth www.blood-isbt-2025.com
2025 AAMRI Convention and Dinner November 4–5, Canberra aamri.org.au/2025-aamri-convention-dinner
AEN 2025 Conference November 6–7, Newcastle arms.eventsair.com/aen-2025-conference
Acoustics 2025 — Sounds of the Sunset Coast November 12–14, Joondalup www.acoustics.org.au/events/acoustics-2025
International Research Integrity Conference November 16–18, Sydney researchintegrityconf.com
AIMOS Conference 2025 November 19–21, Sydney aimos-inc.github.io/aimos.conference.2025
CYTO-Connect Perth November 27–29, Perth cytoconnectperth2025.com.au
AIP Summer Meeting 2025 December 1–5, Wollongong aip-summer-meeting.com
Westwick-Farrow Media
A.B.N. 22 152 305 336 www.wfmedia.com.au
Head Office
Unit 5, 6-8 Byfield Street, (Locked Bag 2226) North Ryde BC NSW 1670, AUSTRALIA Ph: +61 2 9168 2500
Editor Lauren Davis LLS@wfmedia.com.au
Publishing Director/MD Janice Williams
Art Director/Production Manager Linda Klobusiak
Art/Production Marija Tutkovska
Circulation Alex Dalland circulation@wfmedia.com.au
Copy Control Ashna Mehta copy@wfmedia.com.au
Advertising Sales
Sales Manager: Kerrie Robinson Ph:0400 886 311 krobinson@wfmedia.com.au
Andrew Jackson Ph: 0400 604 646 ajackson@wfmedia.com.au
Tim Thompson Ph: 0421 623 958 tthompson@wfmedia.com.au
If you have any queries regarding our
policy please email privacy@wfmedia.com.au