Final Report
Project Title: The Immune Response of Diadema antillarum and Recovery from Caribbean-wide Mass Mortality
Date: October 25, 2016
Project Number: R-31-1-14
Investigators and affiliation: Gregory Beck
Department of Biology, University of Massachusetts - Boston
Dr. John Ebersole
Department of Biology, University of Massachusetts - Boston
Dates covered: February 1, 2014 – August 31, 2016
Summary of Impacts and Accomplishments:
1. Objectives: This project involved interdisciplinary research by a comparative immunologist (Beck) and an ecologist/evolutionary biologist (Ebersole) to uncover basic features of the immunological responses of Diadema and any possible relationship to recovery. Recovery of Diadema has been found to be progressing at a very slow rate, and the potential deficiency in active response to challenge by LPS (a constituent of gramnegative bacteria) has been confirmed to exist in Diadema antillarum-a, but not in other common Caribbean sea urchins.
The main goal of this study was to examine the variation in immune responses of Diadema antillarum-a on St. Croix, USVI. We hoped to answer the following questions: Is there immune variation within or among local Diadema populations; Does strength of immune responses depend on Diadema density; Is variation in immune response related to any geographic feature?
Our studies suggest that there is little genetic variation in immune response among Diadema individuals, and strongly indicate that there is no variation in immune response among different populations on St. Croix. Since our studies have been limited to Diadema populations on St. Croix, it remains possible that there are significant differences in immune responses at greater spatial scales that could entail greater genetic isolation (i.e., among different Caribbean islands).
We have examined Diadema populations around the island of St. Croix, sampling populations from sites that differed considerably in wave exposure, current regime, and distance from shore. Since immune responses did not differ significantly among sites, it is unlikely that these immune responses are related to local geographic features.
In addition, the populations we have sampled have ranged in density over two orders of magnitude, so the lack of significant variation in immune responses among populations indicates that immune responses do not vary with density among populations within this density range However, Diadema densities were much higher in the past, and immune responses of such very dense Diadema populations may be different from what we now observe in smaller populations.
2. Advancement of the Field: So far, small-scale attempts to promote restoration of Diadema thorough restocking have not succeeded, often due to very high – and unexplained – losses over very short periods of time The results of this study on Diadema antillarum’s immunity have implications for the techniques used to foster recovery or restoration. First, the finding of this study that a persistent immune defect is present in Diadema suggests that managers may want to initiate restoration/recovery efforts that are remote from human influence if a persistent immune defect is found, since Diadema were found to be vulnerable to human gut bacteria at the time of the mass mortality.
Second, the finding that the immune defect does not vary among populations and does not depend on population density of Diadema suggests to managers that restoration efforts should focus on high-density restocking. The benefits of improved fertilization during spawning and improved survivorship of early recruits that may find shelter in the spine canopy of aggregated adults that are gained from high density are likely to outweigh any possible depressive effect of crowding on Diadema immune responses.
For example, if the general immune responses of Diadema have an important defect, managers may need to reevaluate the desirability of promoting high-density patches. The desirable effects gained from high density development of highly virulent pathogens that is a common consequence of crowding (especially in crowded groups with compromised immunity).
If anything, the new findings of variation in the Diadema immune system make the situation regarding its immune responses to the epidemic disease that devastated Caribbean populations in 1983/84 and subsequent lack of recovery even more puzzling. Examinations of the Diadema genome that we are conducting in concert with the immunological work should be illuminating. Further results of this study will lead to some new ideas, and suggest new approaches.
3. Problems Encountered: None
4. Research Impacts: The results of this study on Diadema antillarum’s immunity have implications for the practices used to foster restoration. Attempts to promote restoration of Diadema through restocking have not succeeded, often due to very high – and unexplained – losses over very short periods of time The finding of this study that a persistent immune defect is present in Diadema suggests that managers may want to initiate restocking efforts that are remote from human influence if a persistent immune defect is found, since Diadema were found to be vulnerable to human gut bacteria at the time of the mass mortality.
Since the general immune responses of Diadema have an important defect, managers would need to reevaluate the desirability of promoting high-density patches if immune responses are depressed at high densities – which is a common consequence of crowding. The desirable effects gained from high density development of highly virulent pathogens that (especially in crowded groups with compromised immunity)
In addition, the finding that the immune defect does not vary among populations and does not depend on population density of Diadema suggests to managers that restocking efforts should focus on establishing high-density populations The benefits of improved fertilization during spawning and improved survivorship of early recruits that may find shelter in the spine canopy of aggregated adults that are gained from high density are likely to outweigh any possible depressive effect of crowding on Diadema immune responses.
5. Other Important Accomplishments or Products: Johnathon Onufryk (PhD student), jonathan.onufryk.001@umb.edu. Jonathan is a 4th year PhD student who will be involved in surveys of Diadema populations and in collections of individual urchins for immunological analysis, as well as data analysis.
John DeFilippo (PhD student), john.defilippo001@umb.edu, John is a 3rd year PhD student who has extensive training in invertebrate cellular and molecular immunology. He is currently working on the molecular characterization of the Diadema genome.
With this funding, we have been able to start a collaboration with Stacey Williams (Institute for Socio-Ecological Research, Lajas, Puerto Rico). She has a project where she
collects high amounts of Diadema settlers (<1mm in test size), grows them up in the laboratory and transplants them back to reefs when they reach 3 cm in size. She can collect over 1,000 settlers over a four-month period. Her ultimate goal is to see how corals recover in the Diadema enhanced areas. We will study the settler’s immune response.
To date 3 publications have been submitted for review while there are 2 others being prepared. The papers submitted are: Comparison of Phagocytosis in Three Caribbean Sea Urchins (John DeFilippo, John Ebersole and Gregory Beck); Comparative Study of Coelomocytes and Coelomic Fluids of 3 Species of Tropical Sea Urchins (Diadema antillarum, Tripneustes ventricosa, and Echinometra lucunter) From the Caribbean Sea (Gregory Beck, John DeFilippo and John Ebersole); Diadema antillarum on St. Croix, USVI: Current Status and Interactions with Herbivorous Fishes (Jonathan Onufryk, John P Ebersole, Gregory Beck)
All of the sequence data we generate is being deposited to the National Center for Biotechnology Information (NCBI).
6. Source of Matching Funds: The sources of matching funds ($66,117) were from our home university (The University of Massachusetts – Boston). These were in-kind cost share contributions that included salary, fringe benefits, and indirect costs. The sponsor did not change.
7. New Extramural Funds in Addition to Match: We were able to submit a grant to the National Fish and Wildlife Foundation (NFWF) Coral Reef Conservation Fund with Stacey Williams (see #5 above) as a co-investigator. The project title was: Enhancing reef resilience by restoring populations of the long-spined sea urchin, Diadema antillarum-a in St. Croix, USVI and Guánica, Puerto Rico: Assessing the relationship between the recovery of ESA listed coral species and Diadema. The project start date was to be 5/17 and the end date was to be 4/19. The project was not funded, but we are looking for additional grant opportunities to collaborate on.
The project was designed to enhance the recovery of ESA listed coral species by repopulating the reefs with Diadema. We wanted to uncover the relationship between coral restoration and the recovery of Diadema on coral reefs in St. Croix and the Guánica watershed area of Puerto Rico. The project would have aided in the management practices of coral reef resources and functions that could improve conditions on the coral reefs of the Caribbean Sea. The budget request was for $60,000.
8. Breakdown of Time and Effort Attributed to the PI and Co-PI:
Gregory Beck (PI) – Dr. Beck provided 3.5% of his academic (non-summer) time as Matching funds ($7583 + applicable fringe benefits); this time is spent in trip preparation, immunological assay of coelomic fluid, data interpretation, and overall grant oversight. UPR SeaGrant has paid Dr. Beck for 5 days of summer salary ($1062 + applicable fringe benefits) – time spent in St. Croix finding accessible sites with sufficient Diadema, collecting samples of coelomic fluid, and preparing them for later lab assay.
John Ebersole (Co-PI) – Dr. Ebersole provided 2.99% of his academic (non-summer) time as Matching Funds ($8833 + applicable fringe benefits) – time spent in data analysis and interpretation and manuscript preparation. UPR SeaGrant has paid Dr. Ebersole for 3 days of summer salary – time spent in St. Croix finding accessible sites with sufficient Diadema, collecting samples of coelomic fluid, and preparing them for later lab assay.
9. Benefits: There are no such ancillary benefits associated with the project.
Final Report Narrative
Statement of the Problem:
This project involved interdisciplinary research by a comparative immunologist (Beck) and an ecologist/evolutionary biologist (Ebersole) designed to uncover basic features of the immunological responses of Diadema The main goal of the proposed study was to examine the variation in immune responses of Diadema antillarum-a on St. Croix, USVI. The questions we attempted to answer included: Is there immune variation within or among local Diadema populations? Does strength of immune responses depend on Diadema density? Is variation in immune response related to any geographic feature? These are the things that could lead to basic management practices that could improve conditions on the reefs of the Caribbean Sea.
Aim #1: Diadema Populations at Sites on St. Croix May Be Recovering
Rationale: To determine whether immunological deficiency played a role in the mass mortality – and continue to play a role – in the dynamics of Diadema antillarum populations, we looked at populations that are apparently recovering from the mass mortality.
Experimental plan:
We continued to follow methods we have used previously in St. Croix to continue a Diadema monitoring program there that becomes more informative as its history (>20 yrs.) lengthens. We determined relative rates of population recovery in different parts of St. Croix, U.S.V.I., counting and sizing Diadema at each of nine locations circling the island. At each reef site at each sampling time, SCUBA divers counted Diadema in 14 randomly located 50m ´ 2m transects. Diadema test diameters were recorded as small (<40mm; mean size of 1 yr. old Diadema = 48.6 mm), medium (41-60mm) or large (>60mm). Diameters were measured using slates that had been previously marked in 1cm increments. Despite reducing the accuracy of measurements, this method allowed us to estimate the size classes of Diadema in crevices that calipers could not reach.
To determine current population levels of Diadema, SCUBA divers conducted surveys using randomly placed 25x2m transects at four back-reef sites on St. Croix, USVI, from May 30th through June 11th 2013. All fishes moving through an area 1m to either side of the transect line and 1m above the transect line were identified and enumerated by two divers, and the two counts were compared with the greater number of each species recorded.
The number of transects placed at the four sites varied due to conditions and ranged from 10 at Split Cove and Turner Hole to 11 at Rod Bay and 12 at Salt River. Rod Bay, Salt River and Turner Hole are part of a long-term study of Diadema recovery; Split Cove was added in 2013 to increase geographic coverage of that monitoring study.
To assess recovery, new data were graphed with older data (including dates before the mass mortality event) to depict overall change. Linear regression was used to detect density complementarity of Diadema versus herbivorous fishes.
Results
In the areas we have surveyed on St. Croix, densities of D. antillarum are still less than ten percent of those prior to the mass mortality event, despite a nearly ten-fold increase of D. antillarum since the 1983-84 die-off. Since surveys began sixteen years ago, population
densities on St. Croix have shown large variations among sites over time (Figure 1). It has been observed that local abundances of D. antillarum tend to fluctuate at low population densities, and the large fluctuations observed on St. Croix show variation in both time and space suggest patchy dispersion and possible migration into and out of study areas. It seems likely that similar fluctuations in population have been occurring, unobserved, throughout the wider Caribbean since the die-off in 1983.
Figure 1. Densities (log scale) of Diadema antillarum at eight sites on St. Croix, USVI from 1983 to present (TH = Turner Hole, RB = Rod Bay, Tag = Tague Bay, Sol = Solitude Bay, Pow = Pow Point, YC = Yellow Cliff, SR =

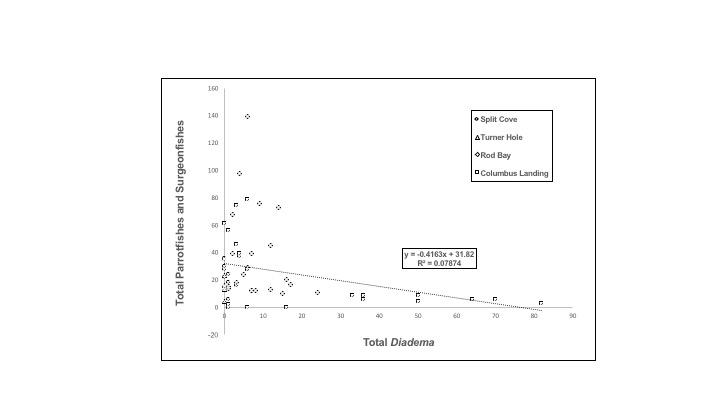
Diadema antillarum continues to interact significantly with roaming herbivorous parrotfishes and surgeonfishes on St. Croix despite the combination of severe reduction in abundance of D. antillarum due to mass mortality and gradual decline in size of herbivorous fishes due to fishing pressures (Hughes et al. 2010). Anecdotally, during our 2013 surveys on St. Croix we rarely observed surgeonfishes larger than 21 centimeters or parrotfishes larger than 36 centimeters, and most were juveniles smaller than 10 cm (pers. obs.), and we saw no raids into damselfish territories by groups of juvenile parrotfishes as seen by Robertson (1976), suggesting that grazing defended damselfish territories may not be worthwhile now that algae are flourishing and competitive pressure is low. Despite the changes in size, abundance, and behavior in both urchins and parrotfishes/surgeonfishes we found that densities of D. antillarum and grazing fishes are inversely related (Figures 2 and 3)
counts (2m radius, 15 minute observation) at Cane Bay on St. Croix, USVI

Data on D. antillarum densities in 2013 contribute to a general picture of irregular and asynchronous fluctuation seen since monitoring began in 2000 (Figure 1). Urchin densities in 2013 densities varied significantly among sites (ANOVA: F = 14.06, df = 3, p = 0.000), with the highest densities at Columbus Landing (0.72 individuals per m2) and the lowest densities at Split Cove (0.02 individuals per m2). At Rod Bay and Columbus Landing, D. antillarum densities increased compared to the most recent prior surveys (in 2010), while in Turner Hole densities declined compared to the most recent survey in that area (in 2008). During the 2016 sampling D. antillarum densities at Columbus Landing were unchanged while densities at Rod Bay increased to an average of 0.7 individuals per m2. While these temporal variations may be considerable it appears that populations in the surveyed areas have recovered, overall, to about 0.1/m2 since the die-off, and despite wide variation from year to year, at no time have all densities dropped back to immediate post-mortality levels. On the other hand, during the last 16 years, overall densities of D. antillarum in the surveyed area, although substantially higher than recorded immediately after the 1983 die-off, are still an order of magnitude below the densities observed prior to the mass mortality event
Numerous herbivorous fishes were found at all sites (36.9 fish m-2 to 63.7 fish m-2), although densities and species composition varied substantially among sites and among transects within sites. A negative association between density of grazing fishes and density of D. antillarum was observed among sites. As expected the relationship between fish and D. antillarum densities were different for roaming grazers compared to the territorials. We found a significant negative relationship between roaming herbivorous fishes and D. antillarum among transects (least squares regression; y = - 0.42x + 31.82, R2 = 0.07, p = 0.033) (Figure 2). Least squares regression analysis of total numbers of densities of territorial herbivorous damselfishes and D. antillarum were not significantly associated (least squares regression; y = - 0.0077x + 9.588, R2 = 0.00006, p = 0.86). Together these analyses indicate that the presence of D. antillarum appears to be negatively affecting roaming grazers, but territorial species appear to be unaffected. Eight stationary point counts again reveal a significant negative association between roaming herbivorous fishes and D. antillarum (least squares regression; y = -0.61x + 27.30, R2 = 0.57, p = 0.030) (Figure 3).
Aim #2: Immune Responses Vary Among Individuals and/or Populations of Diadema
Rationale 1: Since strong immune responses have costs (e.g., auto-immune diseases) as well as benefits, we expect variation in strength of immune response in Diadema antillarum. We characterized the coelomocytes from sea urchins on St. Croix. We focused on variation in the humoral immune response among individuals and populations of D. antillarum. Responses of individuals of other species were used to ensure accuracy and consistency among assays.
Experimental plan 1:
Sea urchins (Diadema antillarum, Tripneustes ventricosa, and Echinometra lucunter) were collected from our study sites around St. Croix and used within 2 hrs of collection. Coelomocytes were isolated from the coelom by piercing the soft tissue surrounding Aristotle’s lantern. We used the coelomocytes in a culture system to assay a key component of the humoral immune response, generation of reactive oxygen intermediates (ROIs) The simple and direct method of detecting this metabolic change that we employed was the NBT reduction assay.
Results 1
Reactive Oxygen Intermediates (ROI) responses of samples of Diadema coelomic fluid taken during all trips to St. Croix (Aug 2016) have been assayed in the lab. Statistical analysis has revealed three interesting findings:
(1) The lack of ROI response to LPS that we first uncovered in 2008 has now been confirmed for all sites we have sampled on St. Croix (see Table 1, below),
(2) Variation in ROI response to LPS among individuals, though statistically meaningful, is not substantial, and
(3) Variation in ROI response to LPS among sites on St. Croix is insubstantial, and probably not significant. ROI assays of coelomic fluid samples taken during our most recent visit to St. Croix have been assayed in the lab and the data are now ready for analysis
The weakness in response to LPS found in preliminary studies has been confirmed. Release of ROI in response to LPS challenge is significantly lower in Diadema than its responses to the standard challenge agents zymosan and peptidoglycan, and significantly lower than the responses to LPS by two other Caribbean sea urchins (E. lucunter and T. ventricosa) as indicated in the 2-way ANOVA table, Table 1, and the Figure 4 below Note that there is only one response failure: Diadema does not respond to LPS.
Table 1 Mean ROI responses of three species of Caribbean sea urchins to three different stimulants, with associated standard deviations and number of individuals sampled. T-statistics and significance levels for the difference between stimulus response and response to control (PBS) *** = p<0.001, ** = p<0.01
Figure 4. Total ROI response of sea urchins (D. antillarum, E. lucunter and T. ventricosa) after exposure to the experimental stimulants. (Error bars = SD).
The ROI response to LPS is not uniformly weak across all Diadema individuals. There are significant differences among Diadema individuals and differences among populations (sites on St. Croix) that are not significant at this point, but may (or may not) rise to significant when the research is complete and sample sizes are larger than we have now. The Model II ANOVA results are in the table below, and the dotgram figure (Figure 5).
Categorical values encountered during processing are: SITE$ (6 levels)
Butler Bay, Cane Bay, Cramer's Par, Gentle Winds, Grassy Point, Sprat Hole
INDIV$ (52 levels)
101.000, 102.000, 103.000, 104.000, 105.000, 106.000, 107.000, 108.000, 109.000, 110.000, 111.000, 112.000, 113.000, 114.000, 115.000, 116.000, 117.000, 118.000, 119.000, 120.000, 121.000, 122.000, 123.000, 124.000, 125.000, 126.000, 127.000, 128.000, 129.000, 130.000, 131.000, 132.000, 133.000, 134.000, 135.000, 136.000, 137.000, 138.000, 139.000, 140.000, 141.000, 142.000, 143.000, 144.000, 145.000, 146.000, 147.000, 148.000, 149.000, 150.000, 151.000, 152.000
REPLICATE$ (3 levels)
A, B, C 4 case(s) deleted due to missing data. Model contains no constant
Dep Var: RESPONSE N: 156 Multiple R: 0.991 Squared multiple R: 0.982
Diadema
Echinometra
Tripneustes
LPS Zymosan
Peptidoglycan
Analysis of Variance:
Variance due to measurement error = 17.6%
Variance due to variation among individuals = 40.2%
Variance due to variation among sites = 42.2%
Figure 5. A dotgram where each point represents the average response, over 3 assays, of a single individual (0 response means no difference from control) from different sites on St. Croix.
ButlerBayCaneBayCramer'sParGentleWindsGrassyPointSpratHole SITE
Rationale 2: We employed next generation sequencing (NGS) to sequence the genome of Echinometra lucunter and compared it to the genome of Diadema antillarum that we have sequenced and to that of the only other sequenced sea urchin, Strongylocentrotus purpuratus. We have identified several gene candidates of the immune system of Diadema that are different from the other species. These comparisons are currently still being investigated. We feel these studies will start to narrow down specific genes (markers) that will be used in further investigations on Diadema immune responses and evolutionary history.
Experimental plan 2a: DNA sequence analysis
Based on a Qubit measurement of quantity, we yielded 4.24 ng/ul (~20nM) of DNA. To
achieve a gDNA library target of 26pM in each reaction we used a dilution factor of 1:769 (20,000pM and divided by 26), and used 20ul for the template reaction. We employed the Template Prep protocol (Ion OneTouch 200 Template Kit v2) and the Sequencing Protocol (Ion PGM 200 Sequencing Kit). Sequence manipulation was done on the Penn Galaxy Server. We uploaded the fastq files to the Galaxy server and ran Galaxy FASTQ Groomer 1.0.4. The filter range 4-37, format fastqsanger was employed. We ran the FASTQ to FASTA tool and converted 6,155,943 FASTQ reads to FASTA. Applying the same filter on Diadema, 90% of bases in a sequence had to have a quality quality cut-off value >= Phred=20 (90% of bases in a sequence had to have a base call accuracy of 90%.), out of 34,736,952 reads, it discarded 22,398,153 (64%) low-quality, leaving 12,338,799 reads.
We filtered sequences by length with a minimal sequence length 175. (For filtering the sequences by length, we arbitrarily deciding to filter out the low tail, and chose a minimal sequence length of 175 bp. Our thinking was, if our sequences were going to be used as queries against a reference genome database, we could get away with smaller reads, but since our sequences were going to be the database that gets queried by Spur's immune gene sequences, the read lengths should probably be longer.
We created a local standalone blast of assorted immune-related genes. We downloaded standalone local BLAST executables from NCBI, ncbi-blast-2.2.26+, and ran makeblastdb to create a blast database of the filtered FASTA file. We also used pdftotext and sort/uniq to extract all SPU gene IDs of immunity-related genes found in the S. purpuratus genome sequences. We ran FASTA grep tool to compare SPU immune gene identifiers against downloaded Sbase fasta, and created peptide fasta of immune genes in each category. W then ran tblasn using peptide.fastas of each gene category as query against blast database of the filtered Diadema and Echinometra FASTA file
Results 2a:
These were the highlights of the blast output.
For D. antillarum
Significant (E >= 0.001) hits to genes in all categories S. purpuratus immune-related genes, except: No significant hits to some of the multiple genes in the following categories:
DD185-333
TNFR-Superfamily
IGV_IGC1-C2_IGSF IL-17
TRAF1-6
TCF-1_LEF-1
Serum-amyloid-protein TOLLIP
For E. lucunter
Significant (E >= 0.001) hits to genes in all categories S. purpuratus immune-related genes, except: No significant hits to some of the multiple genes in the following categories: Amassin B7-like-IGSF Caspase-1-4-5-11-12-13
Cathepsin-A-like
IGV_IGC1-C2_IGSF IKK IL-17
IL-1R_CD121a_&_associated
NLR.9.1
NLR_All
MIF
NLR.1
Perforin-related_(MACPF-domaincontaining)
SARM-related SOCS TIRC
TLR.18
Serum-amyloid-protein
TRAF1-6
TLR.16_Short-TLRs
VEGF xrrc4
For a more complete preliminary analysis, see Appendix. We continue to analyze the sequence data.
Experimental plan 2b:
Transcriptome analysis:
We collected RNA from several of our experimental cultures for use in RNA-Seq analysis. These included samples from D. antillarum, T. ventricosa, and E. lucunter coelomocyte stimulated with seawater (controls) or experimental stimulators (LPS, zymosan, and laminarin). We also collected samples from each of the 3 animals immediately after removal from the water. We have more than 20 samples for analysis.
Results 2b:
We have completed a first run for sequence generation. The initial metrics indicate that the cluster density was fine. This tells us the adapters were ligated successfully and the loading concentration was accurate All 24 of our samples represented. We have performed initial bcl2fastq conversion for fastqc analysis. Fastqc analysis will provide us with visual representation of additional metrics, some indicating how the instrument performed and some providing more insight on the samples themselves (duplication rate estimate, G/C content etc.). We are currently generating alignment data.
Aim #3: The Innate Cellular Response of Diadema Differs from That of Other Caribbean Urchins
Rationale: Since we have identified an apparent lack of response involving secreted defense molecules we wish to see if there is any other fault with the Diadema host defense response. We proposed to study the cellular responses of the sea urchins to look for any Diadema cellular response failings (if any). We used phagocytosis and cytotoxicity assays. We have routinely used all of the following methods in the investigation of host defense responses in invertebrates.
Experimental plan:
We used phagocytosis and cytotoxicity assays to study the cellular immune response of the sea urchins. Phagocytosis [the engulfment of foreign material by specialized cells (i.e., phagocytes)] is the predominant cellular defense mechanism in vertebrates and invertebrates. Phagocytes and cytotoxic cells have been reported in most every invertebrate phylum. Investigators have shown that coelomocytes of the sea urchins were cytotoxic towards allogenic coelomocytes.
Phagocytosis was assessed using microscopy and flow cytometry. Particles (E. coli, yeast, etc.) labeled with pHrodo dye and latex labeled beads were used. A cytotoxicity assay was used as another measure of the strength of the cellular immune response. Cytotoxic activity was assayed by incubating sheep red blood cells as target cells with coelomocytes. The assay was performed with an effector to target cell ratio of from 10 to 1. The effector cells and target cells, or target cells only for control, or target cells only for target spontaneous release, or effector cells only for effector spontaneous release, or medium only for background control were used.
Results:
Phagocytic activity of the coelomocytes was estimated using fluorescently labeled latex beads and microscopy (Figure 6). We used identical view observations between bright filed and fluorescence microscopy. The phagocytosis index that we employed was determined as the percentage of cells that engulfed at least 2 labeled beads. We routinely recorded phagocytosis indexes of > 20 (data not shown). To characterize further the cellular response of the sea urchins we employed flow cytometry to analyze phagocytosis.
Figure Representative fluorescent microscopy of phagocytized fluorescently labeled latex beads in sea urchin coelomocytes stimulated with LPS
scatter (SC) value of 1 were considered margin events and removed. Remaining cells were transformed using an estimateLogicle biexponential function. Cells were plotted for FL1 fluorescent emission (FL1), both with one parameter histograms of channel number vs. number of events using flowViz densityplot, as well as two parameter FL1 vs. SS scatter plots using flowDensity. Density plot gates and cell proportions were partly calculated using deGate. Based on the FL1 density plots, the separation between FL1– and FL1+ cells were determined to be 2.7.
FL1+ cells were those that were positive for phagocytosis of the fluorescently labeled latex beads. Proportions of coelomocytes that were FL1+, as well as cell populations based on granularity and mean FL1 response, were very similar for all urchin species to all treatments (Figure 7 and Tables 2 and 3).
Figure 7. Mean 1D FL1 density plots of D. antillarum, E. lucunter, and T. ventricosus coelomocytes exposed to seawater (A), LPS (B), laminarin (C), and peptidoglycan (D), with the percentage of FL1– and FL1+ cells shown on the cell population peaks.
Table 2. Percentage of FL1+ coelomocytes for three urchin species with four treatments
3. Percentage of phagocytic coelomocytes based on cell granularity
Species
Treatment
–
+
–
With no immune stimulant added (seawater only) Diadema had 0.05% FL1+ cells, Echinometra 0.02%, and Tripneustes 0.03%. With LPS stimulation Diadema had 15% FL1+ cells, Echinometra 14%, and Tripneustes 19%. With laminarin stimulation Diadema had 13% FL1+ cells, Echinometra 16%, and Tripneustes 22%. And with peptidoglycan stimulation Diadema had 10% FL1+ cells, Echinometra 13%, and Tripneustes 14% (Table 2, Figure 7)
To reflect the morphology of phagocytic vs. non-phagocytic cell populations, 2D scatter plots of side-scatter (SS) and FL1 fluorescence were performed, and the percent of cells in each quadrant were determined. The quadrants represent relatively granular nonphagocytic cells, relatively granular phagocytic cells, relatively non-granular nonphagocytic cells, and relatively non-granular non-phagocytic cells. (Table 3).
A one-factor analysis of variance (ANOVA) was performed for each of the three urchin species, with treatment as the independent factor, and the mean FL1+ as the dependent response variable. This showed no statistical difference in any species response to treatments with LPS, laminarin, or peptidoglycan.
We also used a cell cytotoxicity assay to examine the cellular immune response of the urchins. Coelomocytes and RBCs were incubated together for 4 hr at which time the supernatant was isolated and read in a spectrophotometer at an absorbance of 405 nm As shown in Figure 8, cells from all 3 urchin species were capable of lysing the RBCs (an increase in the absorbance). In addition, the cells were stimulated by all experimental treatments.
Table
Diadema Echinometra Tripneustes
Absorbance (OD 405nm)
Positive Control
Discussion:
In these experiments, all urchin species showed a consistent response to all stimulants, as measured by flow cytometry (percent of FL1+ cells) and cytotoxicity. Based on these results it appears that Diadema is capable of microbial recognition, phagocytosis and cytotoxicity. We have previously shown a diminished humoral response by Diadema to LPS, whereas here we find no difference in its cellular response to LPS, with phagocytosis and cytotoxicity of LPS equal to its response to other immune stimulants. While the apparent normal cellular response in Diadema was surprising given our previous findings, more recently proposed models of phagocytosis may offer some possible explanations.
At its most basic level the function of the immune system is host defense: to distinguish self from non-self (i.e., viruses, bacteria, protozoa, fungi, and other non-host proteins) and altered-self (damaged host cells), and to remove pathogens. To accomplish this all gnathostomes (jawed vertebrates) possess both an innate and adaptive immune system. A distinguishing features of adaptive immunity is the somatic rearrangement of genes for its secreted (humoral) antibody molecules, which provides an almost infinite receptor variability to recognize and bind foreign molecules (antigens). Another is the production of antibody-producing memory cells with receptors specific to a previously encountered pathogen and thus able to provide much swifter host defense.
Although true adaptive immunity is confined to the higher vertebrates (variable immune molecules, and alternative anticipatory and memory-like immune mechanisms are found in some invertebrates), all living organisms possess some type of the innate immune system as their first line of defense. For invertebrates to mount a successful immune response, their innate immune system must first be able to first recognize a pathogen, and then take
Figure 8. Mean ± SEM of the absorbance of 405 nm of D. antillarum, T. ventricosus, and E. lucunter phagocytes exposed to seawater, LPS, or zymosan.
D. antillarum T. ventricosus
E. lucunter
Seawater LPS Zymosan
appropriate action to eliminate it by way of humoral and cellular effector mechanisms. Nonself-recognition in invertebrates occurs via germ-line encoded pattern recognition receptors (PRRs), and selective pressure would likely drive an expansion of that repertoire. Indeed, the first sea urchin to be sequenced revealed an amazing complexity and diversity of immune-related genes, making up perhaps 4-5% of the genome.
PRRs include the Toll-like receptors (TLRs), scavenger receptors, peptidoglycan recognition proteins (PGRPs), b-glucan recognition proteins (bGRPs) (also mislabeled as gram-negative binding proteins, GNBPs), cell adhesion receptors such as integrins, and carbohydrate-recognition lectins such as mannose-receptors. Other than Fcg receptors used by vertebrates to bind antibody-opsonized particles, homologs of all PRRs have been found in invertebrates. While their immune function has been shown for some, most others are still to be determined.
These receptors recognize molecular motifs of essential and conserved microbial molecules (pathogen or microbe-associated molecular patterns, PAMPs/MAMPs).
Microbial PAMPs include gram-negative and gram-positive bacterial cell components, fungal b–glucans, and viral nucleic acids However even very similar microbial PAMPs can activate quite different immune responses, and a number of pathogenic bacteria have evolved a variety of ways to avoid these defenses.
Interaction between a receptor and its cognate ligand activates downstream signaling events that lead to various humoral or cellular immune responses, Multiple receptors are involved with ligand binding, and receptor signaling varies by receptor, thus various signaling pathways can be activated. Some of the most studied inflammatory signaling pathways arise from TLRs, and their downstream signal transduction requires engagement of adaptor molecules to activate kinase cascades to transcription factors. Activation of NFkB results in gene transcription of cytokines or invertebrate cytokine-like signal molecules that elicit an inflammatory response.
The few sea urchin genomes that have been sequenced show great diversity in their TLR genes. Compared to 13 mammalian TLR genes, 3 species from the Strongylocentrotidae clade have well over two-hundred, while another belonging to the more basal Toxopneustidae clade has one-quarter the number. Although their cognate ligands and function are still unknown, their enhanced expression in phagocytic coelomocytes suggests a role in immune function. Invertebrate TLRs from a protochordate that were transfected into mammalian cells have been found to activate NF-kB pathways in response to known mammalian ligands. TLR signaling and gene expression can be regulated by non-coding micro RNAs, and a miRNA has been shown to affect phagocytosis of a gram-negative bacteria in coelomocytes of an echinoderm sea cucumber through TLR signaling to NF-kB.
Phagocytosis is the engulfment of large particles (0.5 µm or greater) by specialized cells of the innate immune system. It represents one of the most ancient, highly conserved, and important first line microbial defenses, and is the primary cellular defense for invertebrates. Chemotactic movement of phagocytes towards a site of infection is directed by complement, cytokines, or elements of the pathogen itself. Phagocytosis begins by interaction of microbial ligands with multiple and varied PRRs on phagocytic cells, either directly, or indirectly through opsonin and complement receptors. Opsonins are secreted in response to inflammatory cytokines, and are themselves PRRs, adhering to microbial cells as well activating the complement system for further opsonization of pathogens.
Signaling following phagocyte binding is facilitated by receptor clustering at the binding site. Actin-mediated remodeling of the cytoskeleton enables engulfment and internalization of the particles into a phagosome. The phagosome fuses with a lysosome, where it acquires hydrolase enzymes necessary for particle breakdown, thus creating the mature
phagolysosome. Components of NADPH oxidase are assembled on the phagosomal membrane for production of microbicidal reactive oxygen species (ROS).
In vertebrate phagocytes, microbial engulfment is accompanied by trafficking of membrane TLRs and adapter proteins to the phagosome, where TLR signaling facilitates fusion with lysosomes and phagosome maturation. This may be unique to higher organisms, where phagocytosis also represents a means of antigen presentation and activation of cells of the adaptive immune system. Phagolysosome formation has been shown to occur without TLR signaling from the phagosome, although slower and with impaired bacterial killing. There is still debate as to whether TLR signaling from the phagosome is critical to its maturation, or if it proceeds independently.
While required for inflammatory signaling, TLRs may not function directly as phagocytic receptors. Vertebrate phagocytosis of zymosan and LPS can occur even without the appropriate TLR and adapter proteins, but in their absence gene transcription of inflammatory cytokines does not. TLRs may only be capable of recognizing and binding soluble ligands, with the direct recognition of pathogen surfaces necessary for phagocytosis handled by other phagocyte receptors. For example the active portion of LPS is anchored to the bacterial membrane and not accessible to TLR4. Lipid-A must first be extracted and bound to Lipid Binding Protein (LBP), which passes it to a CD14 co-receptor that is then recognized and bound by a TLR4-MD-2 receptor complex.
In vertebrates it is thought that microbial binding to phagocytes and subsequent ingestion may be dependent on PRRs other than TLRs, while both phagosomal maturation and downstream inflammatory signaling are TLR-dependent. Initiation of phagocytosis and activation of inflammatory signaling may be controlled by separate types of PRRs. True phagocytic receptors, such as mannose and scavenger receptors, trigger actinpolymerization and phagocytosis but cannot activate inflammatory signaling, although they may modulate signaling activated by other PRRs. Other kinds of receptors such as TLRs cannot initiate phagocytosis but can regulate its rate, and their primary function is downstream inflammatory signaling transduction to transcription factors like NF-kB. And other PRRs initiate both phagocytosis and signaling, such as b-glucan recognition Dectin-1. Thus phagocytosis might occur through PRRs, while some defect affecting TLR signaling could result in either inefficient microbicidal action within phagocytes, or failure to produce inflammatory cytokines to stimulate humoral mechanisms, and such a defect would leave the organism susceptible to infection. This could be in one or more TLRs or their adapter molecules.
Research on insects has also challenged the traditional cooperative model of the humoral and cellular arms of the immune response. In this new paradigm, humoral immune activation is seen as a later and secondary response to phagocytosis. As such, phagocytosis might occur independently of any defect in the humoral response.
A possible future area of investigation on the cellular immune response of Diadema might involve further phagocytosis assays using pH sensitive beads to determine whether phagocytosis is followed by phagosomal acidification. Additionally, quantification of mRNA expression patterns of PRRs and their adapter molecules, in response to challenge with microbial ligands at multiple time intervals, would provide insight into signaling cascades involved in the cellular immune response.
APPENDIX
Supplemental Information
Immune-related genes in the Strongylocentrotus purpuratus genome used as a reference genome database (genes categories per SyBase)
I. IMMUNITY
I. A1 Immune receptors - Pattern Recognition Receptors (PRRs)
Toll-like receptors (TLR)
TLR Group IA
Annotated Common Synonyms Ortholog/Homolog Gene Names
SPU_000199 Sp-Tlr003 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_000911 Sp-Tlr008 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_001877 Sp-Tlr010 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_004139 Sp-Tlr019 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_004150 Sp-Tlr020 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_005950 Sp-Tlr030 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_009037 Sp-Tlr050 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_010695 Sp-Tlr058 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_011454 Sp-Tlr060 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_011537 Sp-Tlr062 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_011539 Sp-Tlr063 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_011540 Sp-Tlr064 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_011949 Sp-Tlr067 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_015029 Sp-Tlr079 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_015066 Sp-Tlr080 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_015303 Sp-Tlr082 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_018100 Sp-Tlr091 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_019309 Sp-Tlr103 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_025719 Sp-Tlr140 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_011536 Sp-Tlr061 - Not determined
SPU_015533 Sp-Tlr083 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_018519 Sp-Tlr098 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_019042 Sp-Tlr102 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_025263 Sp-Tlr138 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_025312 Sp-Tlr139 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_001650 Sp-Tlr160 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_009952 Sp-Tlr175 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_011277 Sp-Tlr179 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_011455 Sp-Tlr181 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_030080 Sp-Tlr187 - Not determined
SPU_030094 Sp-Tlr210 - Not determined
SPU_021502 Sp-Tlr212 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_021907 Sp-Tlr213 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_021908 Sp-Tlr214 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_030137 Sp-Tlr215 - Not determined
SPU_030076 Sp-Tlr224 - Not determined
SPU_030079 Sp-Tlr227 - Not determined
SPU_004957 Sp-Tlr024 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_007850 Sp-Tlr040 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_010619 Sp-Tlr057 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_010940 Sp-Tlr059 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_013751 Sp-Tlr073 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_021936 Sp-Tlr114 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_023544 Sp-Tlr121 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_009450 Sp-Tlr141 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_027798 Sp-Tlr151 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_020652 Sp-Tlr206 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_020654 Sp-Tlr207 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_027735 Sp-Tlr220 - Urchin-specific homolog of gene family defined by Toll-like receptor
TLR Group IB
Annotated Common Synonyms Ortholog/Homolog Gene Names
SPU_002538 Sp-Tlr015 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_006164 Sp-Tlr031 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_007105 Sp-Tlr035 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_008962 Sp-Tlr048 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_008963 Sp-Tlr049 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_012464 Sp-Tlr069 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_014041 Sp-Tlr075 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_014191 Sp-Tlr077 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_024868 Sp-Tlr134 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_009933 Sp-Tlr174 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_020258 Sp-Tlr203 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_020644 Sp-Tlr205 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_030092 Sp-Tlr001 - Not determined
SPU_015553 Sp-Tlr192 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_019661 Sp-Tlr201 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_020259 Sp-Tlr105 - Urchin-specific homolog of gene family defined by Toll-like receptor
TLR Group IC
Annotated Common Synonyms Ortholog/Homolog Gene Names
SPU_005850 Sp-Tlr029 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_007429 Sp-Tlr037 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_011570 Sp-Tlr065 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_024204 Sp-Tlr123 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_024205 Sp-Tlr124 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_024208 Sp-Tlr126 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_030093 Sp-Tlr002 - Not determined
SPU_002803 Sp-Tlr163 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_016554 Sp-Tlr195 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_024590 Sp-Tlr218 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_007430 Sp-Tlr038 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_022451 Sp-Tlr115 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_024207 Sp-Tlr125 - Urchin-specific homolog of gene family defined by Toll-like receptor
TLR Group ID
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_016536 Sp-Tlr087 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_028893 Sp-Tlr156 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_000375 Sp-Tlr157 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_018380 Sp-Tlr200 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_025136 Sp-Tlr137 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_003846 Sp-Tlr165 - Not determined
SPU_009459 Sp-Tlr173 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_016388 Sp-Tlr193 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_023491 Sp-Tlr216 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_024501 Sp-Tlr217 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_024960 Sp-Tlr135 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_020741 Sp-Tlr208 - Urchin-specific homolog of gene family defined by Toll-like receptor
TLR Group IE
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_004311 Sp-Tlr021 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_014266 Sp-Tlr078 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_024385 Sp-Tlr127 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_024429 Sp-Tlr130 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_007986 Sp-Tlr042 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_024386 Sp-Tlr128 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_027815 Sp-Tlr152 - Urchin-specific homolog of gene family defined by Toll-like receptor
TLR Group I (Orphan)
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_002442 Sp-Tlr014 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_009173 Sp-Tlr052 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_017529 Sp-Tlr089 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_023321 Sp-Tlr120 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_027721 Sp-Tlr150 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_028576 Sp-Tlr154 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_017530 Sp-Tlr197 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_009829 Sp-Tlr054 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_010693 Sp-Tlr178 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_003684 Sp-Tlr018 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_028404 Sp-Tlr153 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_004655 Sp-Tlr166 - Urchin-specific homolog of gene family defined by Toll-like receptor
TLR Group IIA
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_000871 Sp-Tlr007 - Urchin-specific homolog of gene family defined by Toll-like receptor
SPU_002224 Sp-Tlr013 - Not determined
SPU_005088 Sp-Tlr025 - Not determined
SPU_006458 Sp-Tlr033 - Not determined
SPU_008456 Sp-Tlr047 - Not determined
SPU_013824 Sp-Tlr074 - Not determined
SPU_017180 Sp-Tlr088 - Not determined
SPU_018534 Sp-Tlr099 - Not determined
SPU_021395 Sp-Tlr110 - Not determined
SPU_026200 Sp-Tlr142 - Not determined
SPU_027162 Sp-Tlr145 - Not determined
SPU_027164 Sp-Tlr147 - Not determined
SPU_016501 Sp-Tlr086 - Not determined
SPU_006218 Sp-Tlr032 - Not determined
SPU_027163 Sp-Tlr146 - Not determined
TLR Group IIB
Annotated Common Synonyms
Gene Names
SPU_000428 Sp-Tlr004 - Not determined
SPU_004360 Sp-Tlr022 - Not determined
SPU_004791 Sp-Tlr023 - Not determined
SPU_005339 Sp-Tlr026 - Not determined
SPU_021415 Sp-Tlr111 - Not determined
SPU_024733 Sp-Tlr132 - Not determined
SPU_004792 Sp-Tlr167 - Not determined
SPU_005148 Sp-Tlr169 - Not determined
SPU_007418 Sp-Tlr036 - Not determined
SPU_006939 Sp-Tlr034 - Not determined
SPU_020428 Sp-Tlr204 - Not determined
SPU_021225 Sp-Tlr109 - Not determined
TLR Group IIIA
Annotated Common Synonyms
Gene Names
SPU_000615 Sp-Tlr005 - Not determined
SPU_009435 Sp-Tlr053 - Not determined
SPU_012257 Sp-Tlr068 - Not determined
SPU_016468 Sp-Tlr085 - Not determined
SPU_021162 Sp-Tlr108 - Not determined
SPU_021420 Sp-Tlr112 - Not determined
SPU_023035 Sp-Tlr119 - Not determined
SPU_001862 Sp-Tlr161 - Not determined
SPU_004951 Sp-Tlr168 - Not determined
SPU_008229 Sp-Tlr171 - Not determined
SPU_011481 Sp-Tlr182 - Not determined
SPU_016438 Sp-Tlr194 - Not determined
SPU_021362 Sp-Tlr211 - Not determined
SPU_024847 Sp-Tlr219 - Not determined
Ortholog/Homolog
Ortholog/Homolog
SPU_024479 Sp-Tlr222 - Not determined
SPU_009129 Sp-Tlr051 - Not determined
SPU_025076 Sp-Tlr136 - Not determined
SPU_000986 Sp-Tlr158 - Not determined
SPU_001993 Sp-Tlr162 - Not determined
SPU_003419 Sp-Tlr164 - Not determined
SPU_030136 Sp-Tlr170 - Not determined
SPU_009343 Sp-Tlr172 - Not determined
SPU_015333 Sp-Tlr221 - Not determined
SPU_030075 Sp-Tlr223 - Not determined
SPU_030077 Sp-Tlr225 - Not determined
SPU_030078 Sp-Tlr226 - Not determined
SPU_000985 Sp-Tlr009 - Not determined
SPU_010320 Sp-Tlr176 - Not determined
SPU_014352 Sp-Tlr188 - Not determined
SPU_017794 Sp-Tlr199 - Not determined
SPU_019882 Sp-Tlr202 - Not determined
TLR Group IV
Annotated Common Synonyms
Gene Names
SPU_007790 Sp-Tlr039 - Not determined
SPU_008278 Sp-Tlr045 - Not determined
SPU_008396 Sp-Tlr046 - Not determined
SPU_018409 Sp-Tlr096 - Not determined
SPU_018410 Sp-Tlr097 - Not determined
SPU_024062 Sp-Tlr122 - Not determined
SPU_027698 Sp-Tlr149 - Not determined
SPU_015185 Sp-Tlr191 - Not determined
SPU_013162 Sp-Tlr186 - Not determined
SPU_021075 Sp-Tlr209 - Not determined
SPU_009970 Sp-Tlr055 - Not determined
SPU_015132 Sp-Tlr081 - Not determined
SPU_010680 Sp-Tlr177 - Not determined
SPU_017104 Sp-Tlr196 - Not determined
TLR Group V
Annotated Common Synonyms
Gene Names
SPU_010575 Sp-Tlr056 - Not determined
SPU_016457 Sp-Tlr084 - Not determined
SPU_018928 Sp-Tlr101 - Not determined
SPU_030095 Sp-Tlr183 - Not determined
SPU_013111 Sp-Tlr185 - Not determined
SPU_017735 Sp-Tlr198 - Not determined
SPU_019834 Sp-Tlr104 - Not determined
SPU_026274 Sp-Tlr143 - Not determined
SPU_026275 Sp-Tlr144 - Not determined
Ortholog/Homolog
Ortholog/Homolog
TLR Group VI
Annotated Common Synonyms
Gene Names
SPU_008267 Sp-Tlr044 - Not determined
SPU_013470 Sp-Tlr071 - Not determined
SPU_018211 Sp-Tlr092 - Not determined
SPU_018212 Sp-Tlr093 - Not determined
SPU_022909 Sp-Tlr116 - Not determined
SPU_022911 Sp-Tlr117 - Not determined
SPU_024731 Sp-Tlr131 - Not determined
SPU_018213 Sp-Tlr094 - Not determined
TLR Group VII
Annotated Common Synonyms
Gene Names
SPU_003578 Sp-Tlr016 - Not determined
SPU_003579 Sp-Tlr017 - Not determined
SPU_018055 Sp-Tlr090 - Not determined
SPU_021787 Sp-Tlr113 - Not determined
SPU_011328 Sp-Tlr180 - Not determined
TLR Orphan
Annotated Common Synonyms
Gene Names
SPU_013676 Sp-Tlr072 - Not determined
SPU_014073 Sp-Tlr076 - Not determined
SPU_023033 Sp-Tlr118 - Not determined
SPU_024404 Sp-Tlr129 - Not determined
SPU_024815 Sp-Tlr133 - Not determined
SPU_027222 Sp-Tlr148 - Not determined
SPU_028639 Sp-Tlr155 - Not determined
TLR Fly-like
Annotated Common Synonyms
Gene Names
SPU_007859 Sp-Tlr041 - Not determined
SPU_008228 Sp-Tlr043 - Not determined
SPU_011823 Sp-Tlr066 - Not determined
Short TLRs
Annotated Common Synonyms
Gene Names
SPU_001970 Sp-Tlr011 - Not determined
SPU_001971 Sp-Tlr012 - Not determined
SPU_020996 Sp-Tlr106 - Not determined
SPU_020997 Sp-Tlr107 - Not determined
SPU_001458 Sp-Tlr159 - Not determined
Ortholog/Homolog
Ortholog/Homolog
Ortholog/Homolog
Ortholog/Homolog
Ortholog/Homolog
TLR Intron containing
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_018838 Sp-Tlr100 - Not determined
SPU_005830 Sp-Tlr027 - Not determined
SPU_005832 Sp-Tlr028 - Not determined
Additional TIR-containing
Annotated Common Synonyms
Gene Names
SPU_008510 Sp-Hypp_551 hypothetical protein-551 tolllike receptor-like
SPU_013137 Sp-Hypp_2071 hypothetical protein-2071 tolllike receptor-like
NACHT domain and leucine-rich repeat (NLR) proteins
NLR Cluster 1
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
Likely ortholog of CAR62394.1
Likely ortholog of XP_968875.1
Ortholog/Homolog
SPU_000863 Sp-Nlr152 NACHT and LRR containing protein Not determined
SPU_001549 Sp-Nlr169 NACHT and LRR containing protein Not determined
SPU_001884 Sp-Nlr159 NACHT and LRR containing protein Not determined
SPU_002888 Sp-Nlr172 NACHT and LRR containing protein Not determined
SPU_003247 Sp-Nlr133 NACHT and LRR containing protein Not determined
SPU_003539 Sp-Nlr3 NACHT and LRR containing protein Not determined
SPU_003553 Sp-Nlr2 NACHT and LRR containing protein Not determined
SPU_003797 Sp-Nlr136 NACHT and LRR containing protein Not determined
SPU_005026 Sp-Nlr124 NACHT and LRR containing protein Not determined
SPU_005383 Sp-Nlr140 NACHT and LRR containing protein Not determined
SPU_006610 Sp-Nlr4 NACHT and LRR containing protein Not determined
SPU_007446 Sp-Nlr150 NACHT and LRR containing protein Not determined
SPU_008498 Sp-Nlr134 NACHT and LRR containing protein Not determined
SPU_008707 Sp-Nlr149 NACHT and LRR containing protein Not determined
SPU_009659 Sp-Nlr127 NACHT and LRR containing protein Not determined
SPU_010091 Sp-Nlr176 NACHT and LRR containing protein Not determined
SPU_015033 Sp-Nlr195 NACHT and LRR containing protein Not determined
SPU_011097 Sp-Nlr196 NACHT and LRR containing protein Not determined
SPU_011776 Sp-Nlr44 NACHT and LRR containing protein Not determined
SPU_011855 Sp-Nlr130 NACHT and LRR containing protein Not determined
SPU_013038 Sp-Nlr193 NACHT and LRR containing protein Not determined
SPU_016060 Sp-Nlr192 NACHT and LRR containing protein Not determined
SPU_016257 Sp-Nlr142 NACHT and LRR containing protein Not determined
SPU_016810 Sp-Nlr138_1 NACHT and LRR containing protein Not determined
SPU_016926 Sp-Nlr143 NACHT and LRR containing protein Not determined
SPU_017196 Sp-Nlr132 NACHT and LRR containing protein Not determined
SPU_017505 Sp-Nlr131 NACHT and LRR containing protein Not determined
SPU_018384 Sp-Nlr120 NACHT and LRR containing protein Not determined
SPU_022001 Sp-Nlr137 NACHT and LRR containing protein Not determined
SPU_022564 Sp-Nlr191 NACHT and LRR containing protein Not determined
SPU_023183 Sp-Nlr141 NACHT and LRR containing protein Not determined
SPU_025166 Sp-Nlr126 NACHT and LRR containing protein Not determined
SPU_025167 Sp-Nlr156 NACHT and LRR containing protein Not determined
SPU_025179 Sp-Nlr123 NACHT and LRR containing protein Not determined
SPU_026036 Sp-Nlr151 NACHT and LRR containing protein Not determined
SPU_026400 Sp-Nlr155 NACHT and LRR containing protein Not determined
SPU_026622 Sp-Nlr139 NACHT and LRR containing protein Not determined
SPU_027035 Sp-Nlr194 NACHT and LRR containing protein Not determined
SPU_027207 Sp-Nlr190 NACHT and LRR containing protein Not determined
SPU_027511 Sp-Nlr119 NACHT and LRR containing protein Not determined
SPU_027610 Sp-Nlr135 NACHT and LRR containing protein Not determined
SPU_027808 Sp-Nlr154 NACHT and LRR containing protein Not determined
SPU_028820 Sp-Nlr138 NACHT and LRR containing protein Not determined
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_000672 Sp-Nlr41 NACHT and LRR containing protein Not determined
SPU_001054 Sp-Nlr48 NACHT and LRR containing protein Not determined
SPU_001608 Sp-Nlr181 NACHT and LRR containing protein Not determined
SPU_002423 Sp-Nlr56 NACHT and LRR containing protein Not determined
SPU_003200 Sp-Nlr9 NACHT and LRR containing protein Not determined
SPU_004053 Sp-Nlr13 NACHT and LRR containing protein Not determined
SPU_006219 Sp-Nlr198 NACHT and LRR containing protein Not determined
SPU_008547 Sp-Nlr50 NACHT and LRR containing protein Not determined
SPU_009017 Sp-Nlr10 NACHT and LRR containing protein Not determined
SPU_010053 Sp-Nlr102 NACHT and LRR containing protein Not determined
SPU_013504 Sp-Nlr51 NACHT and LRR containing protein Not determined
SPU_015105 Sp-Nlr63 NACHT and LRR containing protein Not determined
SPU_015768 Sp-Nlr179 NACHT and LRR containing protein Not determined
SPU_015972 Sp-Nlr33 NACHT and LRR containing protein Not determined
SPU_016794 Sp-Nlr114 NACHT and LRR containing protein Not determined
SPU_016921 Sp-Nlr49 NACHT and LRR containing protein Not determined
SPU_017129 Sp-Nrl25 NACHT and LRR containing protein Not determined
SPU_017341 Sp-Nlr52 NACHT and LRR containing protein Not determined
SPU_021447 Sp-Nlr54 NACHT and LRR containing protein Not determined
SPU_025077 Sp-Nlr46 NACHT and LRR containing protein Not determined
SPU_025680 Sp-Nlr7 NACHT and LRR containing protein Not determined
SPU_026921 Sp-Nlr14 NACHT and LRR containing protein Not determined
SPU_027513 Sp-Nlr47 NACHT and LRR containing protein Not determined
SPU_028485 Sp-Nlr182 NACHT and LRR containing protein Not determined
SPU_028681 Sp-Nlr8 NACHT and LRR containing protein Not determined
NLR Cluster 3
Annotated Common Synonyms
Ortholog/Homolog Gene Names
SPU_000457 Sp-Nlr91 NACHT and LRR containing protein Not determined
NLR Cluster 2
SPU_006203 Sp-Nlr93 NACHT and LRR containing protein Not determined
SPU_008283 Sp-Nlr88 NACHT and LRR containing protein Not determined
SPU_009111 Sp-Nlr24 NACHT and LRR containing protein Not determined
SPU_023120 Sp-Nlr79 NACHT and LRR containing protein Not determined
SPU_025204 Sp-Nlr90 NACHT and LRR containing protein Not determined
NLR Cluster 4
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_001548 Sp-Nlr53 NACHT and LRR containing protein Not determined
SPU_001781 Sp-Nlr20 NACHT and LRR containing protein Not determined
SPU_002641 Sp-Nlr12 NACHT and LRR containing protein Not determined
SPU_005462 Sp-Nlr65 NACHT and LRR contaning protein Not determined
SPU_006733 Sp-Nlr86 NACHT and LRR containing protein Not determined
SPU_008597 Sp-Nlr76 NACHT and LRR containing protein Not determined
SPU_017054 Sp-Nlr17 NACHT and LRR containing protein Not determined
SPU_019497 Sp-Nlr185 NACHT and LRR containing protein Not determined
SPU_024649 Sp-Nlr62 NACHT and LRR contaning protein Not determined
NLR Cluster 5
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_002868 Sp-Nlr27 NACHT and LRR containing protein Not determined
SPU_003186 Sp-Nlr72 NACHT and LRR containing protein Not determined
SPU_005410 Sp-Nlr184 NACHT and LRR containing protein Not determined
SPU_006019 Sp-Nlr183 NACHT and LRR containing protein Not determined
SPU_008431 Sp-Nlr61 NACHT and LRR contaning protein Not determined
SPU_013465 Sp-Nlr89 NACHT and LRR containing protein Not determined
SPU_015340 Sp-Nlr16 NACHT and LRR containing protein Not determined
SPU_017993 Sp-Nlr18 NACHT and LRR containing protein Not determined
SPU_020380 Sp-Nlr29 NACHT and LRR containing protein Not determined
SPU_021478 Sp-Nlr58 NACHT and LRR containing protein Not determined
SPU_022394 Sp-Nlr60 NACHT and LRR containing protein Not determined
SPU_022780 Sp-Nlr26 NACHT and LRR containing protein Not determined
SPU_023550 Sp-Nlr67 NACHT and LRR containing protein Not determined
SPU_025138 Sp-Nlr68 NACHT and LRR containing protein Not determined
SPU_026071 Sp-Nlr57 NACHT and LRR containing protein Not determined
SPU_028294 Sp-Nlr42 NACHT and LRR containing protein Not determined
SPU_028387 Sp-Nlr69 NACHT and LRR containing protein Not determined
NLR Cluster 6
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_000015 Sp-Nlr94 NACHT and LRR containing protein Not determined
SPU_000852 Sp-Nlr71 NACHT and LRR containing protein Not determined
SPU_000896 Sp-Nlr28 NACHT and LRR containing protein Not determined
SPU_001423 Sp-Nlr200 NACHT and LRR containing protein Not determined
SPU_001444 Sp-Nlr78 NACHT and LRR containing protein Not determined
SPU_001630 Sp-Nlr166 NACHT and LRR containing protein Not determined
SPU_007620 Sp-Nlr187 NACHT and LRR containing protein Not determined
SPU_008382 Sp-Nlr21 NACHT and LRR containing protein Not determined
SPU_014503 Sp-Nlr74 NACHT and LRR containing protein Not determined
SPU_015481 Sp-Nlr85 NACHT and LRR containing protein Not determined
SPU_017245 Sp-Nlr73 NACHT and LRR containing protein Not determined
SPU_019699 Sp-Nlr82 NACHT and LRR containing protein Not determined
SPU_019700 Sp-Nlr77 NACHT and LRR containing protein Not determined
SPU_020240 Sp-Nlr110 NACHT and LRR containing protein Not determined
SPU_021370 Sp-Nlr186 NACHT and LRR containing protein Not determined
SPU_022130 Sp-Nlr70 NACHT and LRR containing protein Not determined
SPU_023628 Sp-Nlr23 NACHT and LRR containing protein Not determined
SPU_028805 Sp-Nlr75 NACHT and LRR containing protein Not determined
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_000816 Sp-Nlr81 NACHT and LRR containing protein Not determined
SPU_001016 Sp-Nlr113 NACHT and LRR containing protein Not determined
SPU_002272 Sp-Nlr105 NACHT and LRR containing protein Not determined
SPU_002372 Sp-Nlr55 NACHT and LRR containing protein Not determined
SPU_003934 Sp-Nlr64 NACHT and LRR containing protein Not determined
SPU_004043 Sp-Nlr30 NACHT and LRR containing protein Not determined
SPU_004165 Sp-Nlr84 NACHT and LRR containing protein Not determined
SPU_004872 Sp-Nlr19 NACHT and LRR containing protein Not determined
SPU_005993 Sp-Nlr92 NACHT and LRR containing protein Not determined
SPU_006456 Sp-Nlr161 NACHT and LRR containing protein Not determined
SPU_008833 Sp-Nlr112 NACHT and LRR containing protein Not determined
SPU_009488 Sp-Nlr22 NACHT and LRR containing protein Not determined
SPU_014128 Sp-Nlr11 NACHT and LRR containing protein Not determined
SPU_019696 Sp-Nlr95 NACHT and LRR containing protein Not determined
SPU_026020 Sp-Nlr59 NACHT and LRR containing protein Not determined
SPU_026304 Sp-Nlr180 NACHT and LRR containing protein Not determined
SPU_028060 Sp-Nlr122 NACHT and LRR containing protein Not determined
SPU_028433 Sp-Nlr129 NACHT and LRR containing protein Not determined
SPU_028483 Sp-Nlr80 NACHT and LRR containing protein Not determined
SPU_028595 Sp-Nlr115 NACHT and LRR containing protein Not determined
SPU_028630 Sp-Nlr107 NACHT and LRR containing protein Not determined
NLR Cluster 8.1
Annotated Common Synonyms Ortholog/Homolog Gene Names
SPU_005609 Sp-Nlr128 NACHT and LRR containing protein Not determined
SPU_011439 Sp-Nlr103 NACHT and LRR containing protein Not determined
SPU_011441 Sp-Nlr101 NACHT and LRR containing protein Not determined
SPU_017708 Sp-Nlr37 NACHT and LRR containing protein Not determined
SPU_025600 Sp-Nlr147 NACHT and LRR containing protein Not determined
NLR Cluster 7
NLR Cluster 8.2
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_003715 Sp-Nlr38 NACHT and LRR containing protein Not determined
SPU_006229 Sp-Nlr104 NACHT and LRR containing protein Not determined
SPU_010667 Sp-Nlr189 NACHT and LRR containing protein Not determined
SPU_013952 Sp-Nlr83 NACHT and LRR containing protein Not determined
SPU_015052 Sp-Nlr109 NACHT and LRR containing protein Not determined
SPU_017038 Sp-Nlr35 NACHT and LRR containing protein Not determined
SPU_020916 Sp-Nlr1 NACHT and LRR containing protein Not determined
NLR Cluster 8.3
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_002436 Sp-Nlr34 NACHT and LRR containing protein Not determined
SPU_003366 Sp-Nlr118 NACHT and LRR containing protein Not determined
SPU_003640 Sp-Nlr116 NACHT and LRR containing protein Not determined
SPU_005301 Sp-Nlr108 NACHT and LRR containing protein Not determined
SPU_005581 Sp-Nlr87 NACHT and LRR containing protein Not determined
SPU_010097 Sp-Nlr106 NACHT and LRR containing protein Not determined
SPU_013206 Sp-Nlr45 NACHT and LRR containing protein Not determined
SPU_014112 Sp-Nlr32 NACHT and LRR containing protein Not determined
SPU_014761 Sp-Nlr111 NACHT and LRR containing protein Not determined
SPU_021243 Sp-Nlr40 NACHT and LRR containing protein Not determined
SPU_021844 Sp-Nlr100 NACHT and LRR containing protein Not determined
SPU_022294 Sp-Nlr39 NACHT and LRR containing protein Not determined
SPU_024975 Sp-Nlr36 NACHT and LRR containing protein Not determined
SPU_026189 Sp-Nlr121 NACHT and LRR containing protein Not determined
SPU_027858 Sp-Nlr31 NACHT and LRR containing protein Not determined
NLR Cluster 9.1
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_003619 Sp-Nlr201 NACHT and LRR containing protein Not determined
SPU_005732 Sp-Nlr168 NACHT and LRR containing protein Not determined
SPU_011088 Sp-Nlr157 NACHT and LRR containing protein Not determined
SPU_015206 Sp-Nlr160 NACHT and LRR containing protein Not determined
SPU_021930 Sp-Nlr197 NACHT and LRR containing protein Not determined
SPU_022070 Sp-Nlr158 NACHT and LRR containing protein Not determined
SPU_022441 Sp-Nlr163 NACHT and LRR containing protein Not determined
SPU_027300 Sp-Nlr170 NACHT and LRR containing protein Not determined
NLR Cluster 9.2
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_001210 Sp-Nlr144 NACHT and LRR containing protein Not determined
SPU_004343 Sp-Nlr145 NACHT and LRR containing protein Not determined
SPU_011035 Sp-Nlr162 NACHT and LRR containing protein Not determined
SPU_012713 Sp-Nlr167 NACHT and LRR containing protein Not determined
SPU_014122 Sp-Nlr146 NACHT and LRR containing protein Not determined
SPU_023642 Sp-Nlr153 NACHT and LRR containing protein Not determined
SPU_024709 Sp-Nlr173 NACHT and LRR containing protein Not determined
NLR Outliers
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_000523 Sp-Nlr148 NACHT and LRR containing protein Not determined
SPU_002962 Sp-Nlr66 NACHT and LRR containing protein Not determined
SPU_003762 Sp-Nlr117 NACHT and LRR containing protein Not determined
SPU_013619 Sp-Nlr5 NACHT and LRR containing protein Not determined
SPU_023532 Sp-Nlr6 NACHT and LRR containing protein Not determined
SPU_025914 Sp-Nlr15 NACHT and LRR containing protein Not determined
SPU_025948 Sp-Nlr97 NACHT and LRR containing protein Not determined
Scavenger receptor cysteine-rich (SRCR)
SRCR models
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_000337 Sp-Srcr1 - Urchin-specific homolog of gene family defined by SRCR
SPU_000492 Sp-Srcr2 - Urchin-specific homolog of gene family defined by SRCR
SPU_000580 Sp-Srcr3 - Urchin-specific homolog of gene family defined by SRCR
SPU_000646 Sp-Srcr4 - Urchin-specific homolog of gene family defined by SRCR
SPU_000654 Sp-Srcr5 - Urchin-specific homolog of gene family defined by SRCR
SPU_000740 Sp-Srcr6 - Not determined
SPU_000876 Sp-Srcr7 - Not determined
SPU_000984 Sp-Srcr8 - Not determined
SPU_001172 Sp-Srcr11 - Not determined
SPU_001177 Sp-Srcr12 - Not determined
SPU_001229 Sp-Srcr13 - Not determined
SPU_001266 Sp-Srcr14 - Not determined
SPU_001727 Sp-Srcr18 - Not determined
SPU_001763 Sp-Srcr19 - Not determined
SPU_001863 Sp-Srcr20 - Urchin-specific homolog of gene family defined by SRCR
SPU_002028 Sp-Srcr21 - Not determined
SPU_002041 Sp-Srcr22 - Not determined
SPU_002350 Sp-Srcr23 - Not determined
SPU_003127 Sp-Srcr24 - Not determined
SPU_003384 Sp-Srcr25 - Urchin-specific homolog of gene family defined by SRCR
SPU_003526 Sp-Srcr26 - Urchin-specific homolog of gene family defined by SRCR
SPU_003778 Sp-Srcr27 - Urchin-specific homolog of gene family defined by SRCR
SPU_003930 Sp-Srcr28 - Urchin-specific homolog of gene family defined by SRCR
SPU_004011 Sp-Srcr33 - Urchin-specific homolog of gene family defined by SRCR
SPU_004086 Sp-Srcr34 - Urchin-specific homolog of gene family defined by SRCR
SPU_004160 Sp-Srcr37 - Urchin-specific homolog of gene family defined by SRCR
SPU_005000 Sp-Srcr40 - Urchin-specific homolog of gene family defined by SRCR
SPU_005154 Sp-Srcr41 - Urchin-specific homolog of gene family defined by SRCR
SPU_005464 Sp-Srcr43 - Urchin-specific homolog of gene family defined by SRCR
SPU_005556 Sp-Srcr44 - Urchin-specific homolog of gene family defined by SRCR
SPU_006045 Sp-Srcr46 - Urchin-specific homolog of gene family defined by SRCR
SPU_006055 Sp-Srcr47 - Urchin-specific homolog of gene family defined by SRCR
SPU_006254 Sp-Srcr48 - Urchin-specific homolog of gene family defined by SRCR
SPU_006264 Sp-Srcr49 - Urchin-specific homolog of gene family defined by SRCR
SPU_006531 Sp-Srcr50 - Urchin-specific homolog of gene family defined by SRCR
SPU_006731 Sp-Srcr53 - Urchin-specific homolog of gene family defined by SRCR
SPU_007110 Sp-Srcr54 - Urchin-specific homolog of gene family defined by SRCR
SPU_007618 Sp-Srcr55 - Urchin-specific homolog of gene family defined by SRCR
SPU_007660 Sp-Srcr56 - Urchin-specific homolog of gene family defined by SRCR
SPU_007718 Sp-Srcr57 - Urchin-specific homolog of gene family defined by SRCR
SPU_007893 Sp-Srcr60 - Not determined
SPU_008432 Sp-Srcr67 - Urchin-specific homolog of gene family defined by SRCR
SPU_008504 Sp-Srcr68 - Urchin-specific homolog of gene family defined by SRCR
SPU_008514 Sp-Srcr69 - Urchin-specific homolog of gene family defined by SRCR
SPU_008598 Sp-Srcr70 - Urchin-specific homolog of gene family defined by SRCR
SPU_008642 Sp-Srcr71 - Urchin-specific homolog of gene family defined by SRCR
SPU_008836 Sp-Srcr72 - Urchin-specific homolog of gene family defined by SRCR
SPU_008885 Sp-Srcr73 - Urchin-specific homolog of gene family defined by SRCR
SPU_009145 Sp-Srcr74 - Urchin-specific homolog of gene family defined by SRCR
SPU_009354 Sp-Srcr75 - Urchin-specific homolog of gene family defined by SRCR
SPU_009496 Sp-Srcr76 - Urchin-specific homolog of gene family defined by SRCR
SPU_009562 Sp-Srcr77 - Urchin-specific homolog of gene family defined by SRCR
SPU_009753 Sp-Srcr80 - Urchin-specific homolog of gene family defined by SRCR
SPU_010001 Sp-Srcr82 - Urchin-specific homolog of gene family defined by SRCR
SPU_010062 Sp-Srcr83 - Urchin-specific homolog of gene family defined by SRCR
SPU_010232 Sp-Srcr86 - Urchin-specific homolog of gene family defined by SRCR
SPU_010409 Sp-Srcr89 - Urchin-specific homolog of gene family defined by SRCR
SPU_010501 Sp-Srcr90 - Not determined
SPU_010523 Sp-Srcr91 - Urchin-specific homolog of gene family defined by SRCR
SPU_010832 Sp-Srcr92 - Urchin-specific homolog of gene family defined by SRCR
SPU_010909 Sp-Srcr93 - Urchin-specific homolog of gene family defined by SRCR
SPU_010953 Sp-Srcr94 - Urchin-specific homolog of gene family defined by SRCR
SPU_010991 Sp-Srcr95 - Urchin-specific homolog of gene family defined by SRCR
SPU_011101 Sp-Srcr99 - Urchin-specific homolog of gene family defined by SRCR
SPU_011146 Sp-Srcr100 - Urchin-specific homolog of gene family defined by SRCR
SPU_011222 Sp-Srcr101 - Urchin-specific homolog of gene family defined by SRCR
SPU_011752 Sp-Srcr102 - Urchin-specific homolog of gene family defined by SRCR
SPU_011977 Sp-Srcr103 - Urchin-specific homolog of gene family defined by SRCR
SPU_012039 Sp-Srcr104 - Urchin-specific homolog of gene family defined by SRCR
SPU_012159 Sp-Srcr105 - Urchin-specific homolog of gene family defined by SRCR
SPU_012410 Sp-Srcr107 - Urchin-specific homolog of gene family defined by SRCR
SPU_012888 Sp-Srcr108 - Urchin-specific homolog of gene family defined by SRCR
SPU_013650 Sp-Srcr109 - Urchin-specific homolog of gene family defined by SRCR
SPU_013831 Sp-Srcr110 - Urchin-specific homolog of gene family defined by SRCR
SPU_013958 Sp-Srcr111 - Urchin-specific homolog of gene family defined by SRCR
SPU_014079 Sp-Srcr112 - Urchin-specific homolog of gene family defined by SRCR
SPU_014080 Sp-Srcr113 - Urchin-specific homolog of gene family defined by SRCR
SPU_014095 Sp-Srcr114 - Urchin-specific homolog of gene family defined by SRCR
SPU_014602 Sp-Srcr115 - Urchin-specific homolog of gene family defined by SRCR
SPU_014829 Sp-Srcr116 - Urchin-specific homolog of gene family defined by SRCR
SPU_014844 Sp-Srcr117 - Urchin-specific homolog of gene family defined by SRCR
SPU_014859 Sp-Srcr118 - Urchin-specific homolog of gene family defined by SRCR
SPU_014860 Sp-Srcr119 - Urchin-specific homolog of gene family defined by SRCR
SPU_015123 Sp-Srcr123 - Urchin-specific homolog of gene family defined by SRCR
SPU_015325 Sp-Srcr124 - Urchin-specific homolog of gene family defined by SRCR
SPU_015387 Sp-Srcr125 - Urchin-specific homolog of gene family defined by SRCR
SPU_015548 Sp-Srcr127 - Urchin-specific homolog of gene family defined by SRCR
SPU_016195 Sp-Srcr132 - Urchin-specific homolog of gene family defined by SRCR
SPU_016531 Sp-Srcr135 - Urchin-specific homolog of gene family defined by SRCR
SPU_016880 Sp-Srcr136 - Urchin-specific homolog of gene family defined by SRCR
SPU_017127 Sp-Srcr137 - Urchin-specific homolog of gene family defined by SRCR
SPU_017194 Sp-Srcr138 - Urchin-specific homolog of gene family defined by SRCR
SPU_017453 Sp-Srcr139 - Urchin-specific homolog of gene family defined by SRCR
SPU_017933 Sp-Srcr140 - Urchin-specific homolog of gene family defined by SRCR
SPU_018252 Sp-Srcr141 - Urchin-specific homolog of gene family defined by SRCR
SPU_018508 Sp-Srcr144 - Urchin-specific homolog of gene family defined by SRCR
SPU_018737 Sp-Srcr145 - Urchin-specific homolog of gene family defined by SRCR
SPU_018939 Sp-Srcr146 - Urchin-specific homolog of gene family defined by SRCR
SPU_018985 Sp-Srcr147 - Urchin-specific homolog of gene family defined by SRCR
SPU_019241 Sp-Srcr148 - Urchin-specific homolog of gene family defined by SRCR
SPU_019291 Sp-Srcr151 - Urchin-specific homolog of gene family defined by SRCR
SPU_019479 Sp-Srcr154 - Urchin-specific homolog of gene family defined by SRCR
SPU_019826 Sp-Srcr155 - Urchin-specific homolog of gene family defined by SRCR
SPU_020081 Sp-Srcr156 - Urchin-specific homolog of gene family defined by SRCR
SPU_020161 Sp-Srcr157 - Urchin-specific homolog of gene family defined by SRCR
SPU_020822 Sp-Srcr161 - Urchin-specific homolog of gene family defined by SRCR
SPU_020868 Sp-Srcr162 - Urchin-specific homolog of gene family defined by SRCR
SPU_021124 Sp-Srcr163 - Urchin-specific homolog of gene family defined by SRCR
SPU_021348 Sp-Srcr164 - Urchin-specific homolog of gene family defined by SRCR
SPU_021457 Sp-Srcr165 - Urchin-specific homolog of gene family defined by SRCR
SPU_021509 Sp-Srcr166 - Urchin-specific homolog of gene family defined by SRCR
SPU_021782 Sp-Srcr169 - Urchin-specific homolog of gene family defined by SRCR
SPU_021789 Sp-Srcr170 - Urchin-specific homolog of gene family defined by SRCR
SPU_021890 Sp-Srcr171 - Urchin-specific homolog of gene family defined by SRCR
SPU_021987 Sp-Srcr172 - Urchin-specific homolog of gene family defined by SRCR
SPU_021988 Sp-Srcr173 - Urchin-specific homolog of gene family defined by SRCR
SPU_022085 Sp-Srcr175 - Urchin-specific homolog of gene family defined by SRCR
SPU_022339 Sp-Srcr190 - Urchin-specific homolog of gene family defined by SRCR
SPU_022814 Sp-Srcr197 - Urchin-specific homolog of gene family defined by SRCR
SPU_023641 Sp-Srcr198 - Urchin-specific homolog of gene family defined by SRCR
SPU_023677 Sp-Srcr199 - Urchin-specific homolog of gene family defined by SRCR
SPU_023840 Sp-Srcr200 - Urchin-specific homolog of gene family defined by SRCR
SPU_023991 Sp-Srcr201 - Urchin-specific homolog of gene family defined by SRCR
SPU_024084 Sp-Srcr202 - Urchin-specific homolog of gene family defined by SRCR
SPU_024408 Sp-Srcr204 - Urchin-specific homolog of gene family defined by SRCR
SPU_024440 Sp-Srcr205 - Urchin-specific homolog of gene family defined by SRCR
SPU_024487 Sp-Srcr206 - Urchin-specific homolog of gene family defined by SRCR
SPU_025862 Sp-Srcr207 - Urchin-specific homolog of gene family defined by SRCR
SPU_025865 Sp-Srcr208 - Urchin-specific homolog of gene family defined by SRCR
SPU_025968 Sp-Srcr209 - Urchin-specific homolog of gene family defined by SRCR
SPU_025983 Sp-Srcr210 - Urchin-specific homolog of gene family defined by SRCR
SPU_026234 Sp-Srcr211 - Urchin-specific homolog of gene family defined by SRCR
SPU_026241 Sp-Srcr212 - Urchin-specific homolog of gene family defined by SRCR
SPU_026408 Sp-Srcr213 - Urchin-specific homolog of gene family defined by SRCR
SPU_026709 Sp-Srcr214 - Urchin-specific homolog of gene family defined by SRCR
SPU_027037 Sp-Srcr217 - Urchin-specific homolog of gene family defined by SRCR
SPU_027379 Sp-Srcr220 - Urchin-specific homolog of gene family defined by SRCR
SPU_027503 Sp-Srcr221 - Urchin-specific homolog of gene family defined by SRCR
SPU_027619 Sp-Srcr222 - Urchin-specific homolog of gene family defined by SRCR
SPU_028233 Sp-Srcr223 - Urchin-specific homolog of gene family defined by SRCR
SPU_028382 Sp-Srcr224 - Urchin-specific homolog of gene family defined by SRCR
SPU_028612 Sp-Srcr225 - Urchin-specific homolog of gene family defined by SRCR
SPU_028669 Sp-Srcr226 - Urchin-specific homolog of gene family defined by SRCR
SPU_028804 Sp-Srcr228 - Urchin-specific homolog of gene family defined by SRCR
SRCR Contiguous (fragmented?) models
Annotated Common Synonyms
Ortholog/Homolog Gene Names
SPU_001004 Sp-Srcr9 - Not determined
SPU_001005 Sp-Srcr10 - Not determined
SPU_003963 Sp-Srcr29 - Urchin-specific homolog of gene family defined by SRCR
SPU_003964 Sp-Srcr30 - Urchin-specific homolog of gene family defined by SRCR
SPU_003965 Sp-Srcr31 - Urchin-specific homolog of gene family defined by SRCR
SPU_003966 Sp-Srcr32 - Urchin-specific homolog of gene family defined by SRCR
SPU_004100 Sp-Srcr35 - Urchin-specific homolog of gene family defined by SRCR
SPU_004101 Sp-Srcr36 - Urchin-specific homolog of gene family defined by SRCR
SPU_006538 Sp-Srcr51 - Urchin-specific homolog of gene family defined by SRCR
SPU_006539 Sp-Srcr52 - Urchin-specific homolog of gene family defined by SRCR
SPU_007781 Sp-Srcr58 - Urchin-specific homolog of gene family defined by SRCR
SPU_007782 Sp-Srcr59 - Urchin-specific homolog of gene family defined by SRCR
SPU_007894 Sp-Srcr61 - Urchin-specific homolog of gene family defined by SRCR
SPU_007895 Sp-Srcr62 - Urchin-specific homolog of gene family defined by SRCR
SPU_007896 Sp-Srcr63 - Not determined
SPU_007897 Sp-Srcr64 - Urchin-specific homolog of gene family defined by SRCR
SPU_007899 Sp-Srcr65 - Urchin-specific homolog of gene family defined by SRCR
SPU_007900 Sp-Srcr66 - Urchin-specific homolog of gene family defined by SRCR
SPU_009676 Sp-Srcr78 - Urchin-specific homolog of gene family defined by SRCR
SPU_009677 Sp-Srcr79 - Urchin-specific homolog of gene family defined by SRCR
SPU_010226 Sp-Srcr84 - Urchin-specific homolog of gene family defined by SRCR
SPU_010227 Sp-Srcr85 - Urchin-specific homolog of gene family defined by SRCR
SPU_010240 Sp-Srcr87 - Urchin-specific homolog of gene family defined by SRCR
SPU_010241 Sp-Srcr88 - Urchin-specific homolog of gene family defined by SRCR
SPU_010992 Sp-Srcr96 - Urchin-specific homolog of gene family defined by SRCR
SPU_010993 Sp-Srcr97 - Urchin-specific homolog of gene family defined by SRCR
SPU_010994 Sp-Srcr98 - Urchin-specific homolog of gene family defined by SRCR
SPU_014992 Sp-Srcr120 - Urchin-specific homolog of gene family defined by SRCR
SPU_014993 Sp-Srcr121 - Urchin-specific homolog of gene family defined by SRCR
SPU_014994 Sp-Srcr122 - Urchin-specific homolog of gene family defined by SRCR
SPU_015937 Sp-Srcr128 - Urchin-specific homolog of gene family defined by SRCR
SPU_015938 Sp-Srcr129 - Urchin-specific homolog of gene family defined by SRCR
SPU_015989 Sp-Srcr130 - Urchin-specific homolog of gene family defined by SRCR
SPU_015991 Sp-Srcr131 - Urchin-specific homolog of gene family defined by SRCR
SPU_016373 Sp-Srcr133 - Urchin-specific homolog of gene family defined by SRCR
SPU_016374 Sp-Srcr134 - Urchin-specific homolog of gene family defined by SRCR
SPU_018429 Sp-Srcr142 - Urchin-specific homolog of gene family defined by SRCR
SPU_018430 Sp-Srcr143 - Urchin-specific homolog of gene family defined by SRCR
SPU_019262 Sp-Srcr149 - Urchin-specific homolog of gene family defined by SRCR
SPU_019263 Sp-Srcr150 - Urchin-specific homolog of gene family defined by SRCR
SPU_021691 Sp-Srcr167 - Urchin-specific homolog of gene family defined by SRCR
SPU_021692 Sp-Srcr168 - Urchin-specific homolog of gene family defined by SRCR
SPU_022145 Sp-Srcr176 - Urchin-specific homolog of gene family defined by SRCR
SPU_022146 Sp-Srcr177 - Urchin-specific homolog of gene family defined by SRCR
SPU_022147 Sp-Srcr178 - Urchin-specific homolog of gene family defined by SRCR
SPU_022148 Sp-Srcr179 - Urchin-specific homolog of gene family defined by SRCR
SPU_022149 Sp-Srcr180 - Urchin-specific homolog of gene family defined by SRCR
SPU_022150 Sp-Srcr181 - Urchin-specific homolog of gene family defined by SRCR
SPU_022151 Sp-Srcr182 - Urchin-specific homolog of gene family defined by SRCR
SPU_022285 Sp-Srcr185 - Urchin-specific homolog of gene family defined by SRCR
SPU_022286 Sp-Srcr186 - Not determined
SPU_022287 Sp-Srcr187 - Urchin-specific homolog of gene family defined by SRCR
SPU_022288 Sp-Srcr188 - Urchin-specific homolog of gene family defined by SRCR
SPU_022289 Sp-Srcr189 - Urchin-specific homolog of gene family defined by SRCR
SPU_022423 Sp-Srcr191 - Urchin-specific homolog of gene family defined by SRCR
SPU_022424 Sp-Srcr192 - Urchin-specific homolog of gene family defined by SRCR
SPU_022567 Sp-Srcr194 - Urchin-specific homolog of gene family defined by SRCR
SPU_022568 Sp-Srcr195 - Urchin-specific homolog of gene family defined by SRCR
SPU_022569 Sp-Srcr196 - Urchin-specific homolog of gene family defined by SRCR
SPU_026848 Sp-Srcr215 - Urchin-specific homolog of gene family defined by SRCR
SPU_026849 Sp-Srcr216 - Urchin-specific homolog of gene family defined by SRCR
SPU_027287 Sp-Srcr218 - Urchin-specific homolog of gene family defined by SRCR
SPU_027288 Sp-Srcr219 - Urchin-specific homolog of gene family defined by SRCR
SRCR and additional domains
Annotated Common Synonyms Ortholog/Homolog Gene Names
SPU_001601 Sp-Srcr16 - Urchin-specific homolog of gene family defined by SRCR
SPU_004642 Sp-Srcr38 - Urchin-specific homolog of gene family defined by SRCR
SPU_005420 Sp-Srcr42 - Urchin-specific homolog of gene family defined by SRCR
SPU_007349 Sp-Srcr39 - Urchin-specific homolog of gene family defined by SRCR
SPU_007370 Sp-Srcr17 - Urchin-specific homolog of gene family defined by SRCR
SPU_007372 Sp-Srcr15 - Urchin-specific homolog of gene family defined by SRCR
SPU_007840 Sp-Srcr45 - Urchin-specific homolog of gene family defined by SRCR
SPU_009220 Sp-Srcr184 - Urchin-specific homolog of gene family defined by SRCR
SPU_010330 Sp-Srcr183 - Urchin-specific homolog of gene family defined by SRCR
SPU_012230 Sp-Srcr106 - Not determined
SPU_015539 Sp-Srcr126 - Urchin-specific homolog of gene family defined by SRCR
SPU_019370 Sp-Srcr152 - Urchin-specific homolog of gene family defined by SRCR
SPU_019374 Sp-Srcr153 - Urchin-specific homolog of gene family defined by SRCR
SPU_020273 Sp-Srcr158 - Urchin-specific homolog of gene family defined by SRCR
SPU_020597 Sp-Srcr159 - Urchin-specific homolog of gene family defined by SRCR
SPU_020650 Sp-Srcr160 - Urchin-specific homolog of gene family defined by SRCR
SPU_022000 Sp-Srcr174 - Urchin-specific homolog of gene family defined by SRCR
SPU_022528 Sp-Srcr193 - Urchin-specific homolog of gene family defined by SRCR
SPU_024390 Sp-Srcr203 - Urchin-specific homolog of gene family defined by SRCR
SPU_028680 Sp-Srcr227 - Urchin-specific homolog of gene family defined by SRCR
SPU_005860 Sp-Srcr/Ptpc1 - Urchin-specific homolog of gene family defined by SRCR
SPU_006659 Sp-Srcr/Lrr1 - Not determined
SPU_009988 Sp-Ig/Srcr1 - Not determined
SPU_009989 Sp-Srcr81 - Urchin-specific homolog of gene family defined by SRCR
CD36-like
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_007495 Sp-Cd36antigenL - Urchin-specific homolog of gene family defined by AAH61853
SPU_003486 Sp-ScavrBIIIL - Urchin-specific homolog of gene family defined by AAQ08185
SPU_018904 Sp-ScavrB1L - Urchin-specific homolog of gene family defined by NP_058021
SPU_020799 Sp-ScavrB2L1 - Urchin-specific homolog of gene family defined by NP_005497
SPU_023671 Sp-Scavr - Urchin-specific homolog of gene subfamily defined by SR repeats
SPU_025565 Sp-ScavrB2L2 - Urchin-specific homolog of gene family defined by NP_001016557
SPU_026720 Sp-ScavrBIIIL_1 - Not determined
Peptidoglycan recognition proteins (PGRP)
Annotated Common Synonyms
Gene Names
SPU_000222 Sp-Pgrp3 - Unclear
SPU_003882 Sp-Pgrp4 - Unclear
Ortholog/Homolog
SPU_007946 Sp-Pgrp2 - Likely ortholog of PGRP-LC
SPU_023247 Sp-Pgrp1 - Likely ortholog of PGRP-SC1b
SPU_030064 Sp-Pgrp5 - Likely ortholog of PGRP-SC2
Gram-negative binding proteins (GNBP)
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_001352 Sp-Ggnbp2 gametogenetin binding protein 2 Likely ortholog of gametogenetin binding protein 2
SPU_006529 Sp-Gnbp1/2/3B - Not determined
SPU_006530 Sp-Gnbp1/2/3B_1 - Not determined
SPU_016163 Sp-Gnbp1/2/3A - Likely ortholog of GNBP1/2/3
SPU_024075 Sp-Gnbp1/2/3C - Not determined
RigIL/LGP2/MDA-5-like
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_000006 Sp-RigIL4 Sp-MDA-5 like 12, Sp-LGP2 like 12 Not determined
SPU_005476 Sp-RigIL12 Sp-MDA-5 like 8, Sp-LGP2 like 8 Not determined
SPU_007126 Sp-RigIL8 Sp-MDA-5 like 5, Sp-LGP2 like 5 Not determined
SPU_010536 Sp-RigIL11 Sp-MDA-5 like 7, Sp-LGP2 like 7 Not determined
SPU_011866 Sp-RigIL6 Sp-MDA-5 like 3, Sp-LGP2 like 3 Not determined
SPU_014118 Sp-RigIL10 Sp-MDA-5 like 6, Sp-LGP2 like 6 Not determined
SPU_014119 Sp-RigIL9 Sp-MDA-5 like 6, Sp-LGP2 like 6 Not determined
SPU_014310 Sp-RigIL1 Sp-MDA-5 like 1, Sp-LGP2 like 1 Not determined
SPU_014311 Sp-RigIL7 Sp-MDA-5 like 4, Sp-LGP2 like 4 Not determined
SPU_016718 Sp-RigIL2 Sp-MDA-5 like 10, Sp-LGP2 like 10 Not determined
SPU_019617 Sp-RigIL13 Sp-MDA-5 like 9, Sp-LGP2 like 9 Not determined
SPU_020020 Sp-RigIL3 Sp-MDA-5 like 11, Sp-LGP2 like 11 Not determined
SPU_025885 Sp-RigIL5 Sp-MDA-5 like 2, Sp-LGP2 like 2 Not determined
SPU_028689 Sp-Lrig1 leucine-rich repeats and immunoglobulinlike domains 1 Likely ortholog of AAU44786.1
IMCV (Ips-1/MAVS/Cardif/VISA)
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_017445 Sp-ImcvL Sp-IPS-1/MAVS/Cardif/VISA like Not determined
I. A2 Immune receptors - C-type lectin domain proteins
Echinoidin-like
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_000839 Sp-EchL1 - Not determined
SPU_000906 Sp-EchL2 - Not determined
SPU_001821 Sp-EchL3 - Not determined
SPU_005127 Sp-EchL4 - Not determined
SPU_015211 Sp-Clect_34 - Unclear
SPU_022861 Sp-Clect_43 - Unclear
SPU_025892 Sp-Clect_52 - Unclear
SPU_008393 Sp-Clect_19 - Unclear
SPU_022396 Sp-Clect_42 - Unclear
SPU_007040 Sp-Clect_16 - Unclear
SPU_007882 Sp-C-lectin - Not determined
Additional small C-type lectin domain proteins
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_028432 Sp-Clect_56 - Unclear
SPU_010101 Sp-Clect_22 - Unclear
SPU_003618 Sp-Clect_8 - Unclear
SPU_014221 Sp-El1L3 secreted lectin homolog Urchin homolog of subfamily defined by HeEL1
SPU_003774 Sp-Clect_9 - Unclear
I. A3 Immune receptors - Ig superfamily
IG V - IG C1/C2 IGSF genes
Annotated Common Synonyms Ortholog/Homolog Gene Names
SPU_013709 Sp-Vc1_1 - Not determined
SPU_030071 Sp-Vc1_2 - Not determined
SPU_030081 Sp-Vc1_3 - Not determined
SPU_000577 Sp-Igvf - Not determined
SPU_011076 Sp-JamL1 Sp-Junctional Adhesion Molecule-like Not determined
SPU_002608 Sp-Vc2_1 - Not determined
SPU_012388 Sp-Igvc1 Sp-IGv-containing Not determined
SPU_024439 Sp-Sirpb/GL1 - Not determined
SPU_024787 Sp-Igvf2 - Not determined
B7-like IGSF genes
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_010746 Sp-B7L1 - Not determined
SPU_017228 Sp-B7L2 - Not determined
SPU_020457 Sp-B7L3 - Not determined
SPU_028300 Sp-B7L4 - Not determined
SPU_028510 Sp-B7L5 - Not determined
ITAM-containing Ig
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_013624 Sp-Ig/Itamc1 Sp-IG/ITAM-containing Not determined
I.A4
Genes associated with VDJ recombination
TdT/Pol-μ
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_009980 Sp-TdTl/Pol/Mu terminal deoxyribonucleotidyltransferase-like Likely ortholog of TdT and POLM
Rag1
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_027600 Sp-Rag1l - Urchin-specific homolog of gene family defined by Rag1
Rag2
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_030091 Sp-Rag2l - Urchin-specific homolog of gene family defined by Rag2
Ku70
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_009797 Sp-Ku70l - Not determined
Ku80
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_009224 Sp-Ku86l - Not determined
SPU_006396 Sp-Bmp5-8 - Not determined
DNA ligase IV
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_018243 Sp-Dnl4 ligase IV, DNA, ATP-dependent Not determined
xrrc4
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_024517 Sp-Xrcc4 - Likely ortholog of Xrcc4, X -ray repair cross complementing protein 4
Xlf / CERNUNNOS
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_012239 Sp-Cern - Likely ortholog of Cernunnos / Xlf
DNA-PKcs-like
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_015484 Sp-Dna_pkcs_1 DNA activated protein kinase Not determined
SPU_015529 Sp-Dna_pkcs DNA activated protein kinase Likely ortholog of DNA-PKcs, catalytic subunit of DNA dependent protein kinase
DNA cross-link repair 1 / Artemis
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_003308 Sp-Dclr1 DNA cross-link repair 1C protein, Artemis protein, PSO2 homolog S.cerevisiae. Not determined
I.B1 Effector Genes - Complement
C3/4/5
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_005182 Sp-064 Sp-C3 Urchin-specific homolog of gene family defined by Complement C3
SPU_012439 Sp-064_1 Sp-C3 Urchin-specific homolog of gene family defined by Complement C3
SPU_017239 Sp-064_3 Sp-C3 Urchin-specific homolog of gene family defined by Complement C3
SPU_018503 Sp-064_2 Sp-C3 Urchin-specific homolog of gene family defined by Complement C3
SPU_000997 Sp-C3-2 - Likely ortholog of Sp-C3
Thioester-containing
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_022988 Sp-Tcp1 thioester containing protein 1 Likely ortholog of Sp-C3 and alpha 2 macroglobulin
SPU_005193 Sp-Tcp2 thioester containing protein 2 Not determined
Mannose Binding Protein
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_004869 Sp-Sm30F - Not determined
C2/Factor B
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_028188 Sp-Fb SpBf Likely ortholog of complement factor B
SPU_028187 Sp-Fb2 SpBf-2 Likely ortholog of complement factor B
SPU_009091 Sp-Fb3 SpBf-3 Likely ortholog of complement factor B
C1q
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_005500 Sp-C1qL - Not determined
SPU_006578 Sp-C1qL_1 - Not determined
SPU_009020 Sp-C1qL_2 - Not determined
SPU_009401 Sp-C1qc - Not determined
CD59
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_030142 Sp-Cd59/Sca2L1 - Not determined
SPU_030143 Sp-Cd59/Sca2L2 - Not determined
Additional thioester containing genes
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_006406 Sp-Tecp1 - Not determined
SPU_026313 - - Not determined
SPU_019422 Sp-Tecp2 - Urchin-specific homolog of gene family defined by thioester site
SPU_013170 Sp-Tecp4 - Urchin-specific homolog of gene family defined by alpha 2 macroglobulin
SPU_013169 Sp-Tecp5 - Urchin-specific homolog of gene family defined by thioester site
I.B2
Effector Genes -
Cytolytic pathways
Perforin-related (MACPF domain containing genes)
Annotated Common Synonyms Ortholog/Homolog Gene Names
SPU_000751 Sp-MacpfD1 - Not determined
SPU_001794 Sp-MacpfC1 - Not determined
SPU_001797 Sp-MacpfC.2 - Not determined
SPU_002548 Sp-MacpfB.0_1 - Not determined
SPU_002549 Sp-MacpfB.0 - Not determined
SPU_002550 Sp-MacpfB.1 - Not determined
SPU_005223 Sp-MacpfA1 - Not determined
SPU_006818 Sp-MacpfG1 - Not determined
SPU_007159 Sp-MacpfE1 - Not determined
SPU_008485 Sp-MacpfE.4 - Not determined
SPU_014229 Sp-MacpfF.1 - Not determined
SPU_014677 Sp-MacpfC3 - Not determined
SPU_014984 Sp-MacpfA.3 - Not determined
SPU_015144 Sp-MacpfB.2 - Not determined
SPU_016546 Sp-MacpfB3 - Not determined
SPU_017952 Sp-MacpfA4 - Not determined
SPU_022091 Sp-MacpfA2 - Not determined
SPU_022230 Sp-MacpfE.3 - Not determined
SPU_022318 Sp-MacpfD.2 - Not determined
SPU_026119 Sp-MacpfD.3 - Not determined
SPU_027405 Sp-MacpfD.4 - Not determined
SPU_028756 Sp-MacpfE.2 Not determined
Cathepsin A-like
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_012846 Sp-Ppgb
SPU_013245 Sp-Ppgb_1
SPU_015939 Sp-Ppgb_3
SPU_026832 Sp-Ppgb_2
Cathepsin B
Lysosomal protective protein precursor (Cathepsin A)
(Carboxypeptidase C) (Protective protein for beta-galactosidase)
[Contains: Lysosomal protective protein 32 kDa chain; Lysosomal protective protein 20 kDa chain]
Lysosomal protective protein precursor (Cathepsin A) (Carboxypeptidase C) (Protective protein for beta-galactosidase)
[Contains: Lysosomal protective protein 32 kDa chain; Lysosomal protective protein 20 kDa chain]
Lysosomal protective protein precursor (Cathepsin A) (Carboxypeptidase C) (Protective protein for beta-galactosidase)
Lysosomal protective protein precursor (Cathepsin A) (Carboxypeptidase C) (Protective protein for beta-galactosidase)
[Contains: Lysosomal protective protein 32 kDa chain; Lysosomal protective protein 20 kDa chain]
Annotated Common Synonyms Ortholog/Homolog Gene Names
SPU_007151 Sp-Ctsb Cathepsin B Not determined
Cathepsin C
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_005834 Sp-Cts1 Sp-Cathepsin1, Sp-CtsC, Sp-CathepsinC Not determined
Cathepsin F
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_014914 Sp-Cts10 Sp-Cathepsin10, Sp-CtsF-l1, Sp-CathepsinF-like1 Not determined
Cathepsin L
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_009042 Sp-Cts2 Sp-Cathepsin2, Sp-CtsL-l1, Sp-CathepsinL-like1 Not determined
SPU_014765 Sp-Cts9 Sp-Cathepsin9, Sp-CtsL-l5, Sp-CathepsinL-like5 Not determined
SPU_014766 Sp-Cthpsn6 Sp-CathepsinL-Like2 Not determined
SPU_014767 Sp-Cts7 Sp-Cathepsin7, Sp-CtsL-l3, Sp-CathepsinL-like3 Not determined
SPU_014768 Sp-Cts8 Sp-Cathepsin8, Sp-CtsL-l4, Sp-CathepsinL-like4 Not determined
SPU_015668 Sp-Cts11 Sp-Cathepsin11, Sp-CtsL-l6, Sp-CathepsinL-like6 Not determined
SPU_020837 Sp-Cts12 Sp-Cathepsin12, Sp-CtsL-l7, Sp-CathepsinL-like7 Not determined
SPU_020838 Sp-Cts13 Sp-Cathepsin13, Sp-CtsL-l8, Sp-CathepsinL-like8 Not determined
Cathepsin Z
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_009601 Sp-Cts4 Sp-Cathepsin4, Sp-CtsZ-l1, Sp-CathepsinZ-like1 Not determined
SPU_013893 Sp-Cts5 Sp-Cathepsin5, Sp-CtsZ-l2, Sp-CathepsinZ-like2 Not determined
Novel Cathepsin
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_009368 Sp-Cts3 Sp-Cathepsin3 Not determined
Granzymes
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_001588 Sp-GramarL2 - Not determined
SPU_016107 Sp-GramarL3 - Not determined
SPU_023706 Sp-GramarL1 - Not determined
I.B3 Effector genes - Additional cytotoxic effectors and regulators
NOS
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_002328 Sp-Nos_1 - Not determined
SPU_013373 Sp-Nos_3 - Not determined
SPU_016283 Sp-Nos_2 - Not determined
SPU_019970 Sp-Nos1 Nitric-oxide synthase, brain (NOS type I) (Neuronal NOS) (N-NOS) (nNOS) (Constitutive NOS) (NC -NOS) (BNOS) Not determined
SPU_025118 Sp-Nos2L nitric oxide synthase 2 [Lehmannia valentiana]-like Likely ortholog of BAH70357.1
Peroxidase
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_008911 Sp-Hdpx1 Sp-Heme-dependent Peroxidase-1 Not determined
SPU_019097 Sp-Hdpx2 Sp-Heme-dependent Peroxidase-2 Not determined
SPU_002004 Sp-Hdpx3 Sp-Heme-dependent Peroxidase-3 Not determined
I.C Sea urchin immune response genes
DD185/333
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_011836 Sp-185/333Ex - Not determined
SPU_019327 Sp-185/333B3d
Not determined
SPU_022178 Sp-185/333E2 - Not determined
SPU_022179 Sp-185/333D1
SPU_030144 Sp-185/333X
Not determined
Not determined
SPU_030145 Sp-185/333C4 - Not determined
SPU_030146 Sp-185/333X_1
SPU_030262 Sp-185/333/E2
SPU_030264 Sp-185/333/D1
II. REGULATORY PATHWAYS
II.A1 Intracellular signal transduction - NFκB / IRF pathways
NFκB
Not determined
Not determined
Not determined
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_008177 Sp-Nfkb - Urchin-specific homolog of gene subfamily defined by gene_is=NFkB_protein_[Strongylocentrotus_purpuratus]
SPU_012203 Sp-Rel - Likely ortholog of C-rel [danio rerio]
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_011197 Sp-IkB Sp-IkappaB, Sp-NF-kappaB Inhibitor Not determined
IKK
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_007638 Sp-Ikke IKK epsilon, inhibitor of nuclear factor kappa B kinase epsilon Not determined
SPU_008254 Sp-Ikk1_1 inhibitor of nuclear factor kappa B kinase 1, IKKalpha Not determined
SPU_008255 Sp-Ikk2 inhibitor of nuclear factor kappa B kinase 2, IKK beta Not determined
SPU_016839 Sp-Ikk1 Inhibitor of nuclear factor kappa B kinase 1, IKKalpha Not determined
SPU_027909 Sp-Ikke_1 inhibitor of nuclear factor kappa B kinase epsilon Not determined
TBK1/NAK
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_004671 Sp-Tbk1 TANK binding kinase 1, NF-kappaB activating kinase, nuclear factor kappa B activating kinase Not determined
TRAF1-6
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_003462 Sp-TrafA - Not determined
SPU_004056 Sp-Trafd1 TRAF-type zinc finger domain containing 1 Likely ortholog of XP_534682.2
SPU_008332 Sp-Traf4 CART1 Likely ortholog of TRAF4
SPU_010527 Sp-Traf - Not determined
SPU_012840 Sp-Traf1_1 - Not determined
SPU_018953 Sp-Traf3_1 TNF receptor-associated factor 3-1 Urchin-specific homolog of gene subfamily defined by NP_663778.1
SPU_019927 Sp-Traf3ip1 TNF receptor-associated factor 3 interacting protein 1 Likely ortholog of XP_001234447.1
SPU_023069 Sp-TrafB - Not determined
SPU_026479 Sp-Traf1 Tnf receptor-associated factor 1 Not determined
SPU_026495 Sp-Traf3 LAP1; CAP-1; CRAF1 Likely ortholog of TRAF3
SPU_028898 Sp-Traf6 - Likely ortholog of Traf6
TRIAD
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_030065 Sp-Triad_2 UBCE7IP1 Not determined
TAK1/MAP3K7
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_002696 Sp-Tak1 TAK1; Mkkk7 Not determined
MAP3K7IP1/TAB1
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_005254 Sp-Tab1 Map3k7ip1 Not determined
TAB2/3
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_003955 Sp-Tab2/3 Map3k7ip2/3 Not determined
UBC13
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_018598 Sp-Ubc13 UBE2N Not determined
UEV1A
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_000742 Sp-Ube2v1/2 Uev1a Not determined
SINK
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_013349 Sp-Shpk SINK-homologous protein kinase; kinase inhibitor of NF-kappaB and p53-mediated transcription Not determined
IRF
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_010404 Sp-Irf - Urchin-specific homolog of gene subfamily defined by interferon_regulatory_factor_1_[Gallus_gallus]
SPU_026877 Sp-Irf4 - Urchin-specific homolog of gene subfamily defined by interferon_regulatory_factor_4_[Gallus_gallus]
II.A2 Intracellular signal transduction - TLR adaptor molecules
MyD88
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_007342 Sp-Myd88_1 - Not determined
SPU_007343 Sp-Myd88 - Not determined
MyD88-Like
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_001905 Sp-Myd88L1 myeloid differentiation primary response gene (88) Not determined
SPU_022707 Sp-Myd88L2 myeloid differentiation primary response gene (88) Not determined
SPU_022708 Sp-Myd88L3 myeloid differentiation primary response gene (88) Not determined
SARM
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_011042 Sp-Sarm - Not determined
Novel family of SARM -related genes
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_000764 Sp-Sarmr4 Sp-Sarm-related 4 Not determined
SPU_003495 Sp-Sarmr10 Sp-Sarm-related 10 Not determined
SPU_004107 Sp-Sarmr12 Sp-Sarm-related 12 Not determined
SPU_004557 Sp-Sarmr11 Sp-Sarm-related 11 Not determined
SPU_006122 Sp-Sarmr1 Sp-Sarm-related 1 Not determined
SPU_007020 Sp-Sarmr3 Sp-Sarm-related 3 Not determined
SPU_007088 Sp-Sarmr14 Sp-Sarm-related 14 Not determined
SPU_008302 Sp-Sarmr13 Sp-Sarm-related 13 Not determined
SPU_015127 Sp-Sarmr5 Sp-Sarm-related 5 Not determined
SPU_018168 Sp-Sarmr9 Sp-Sarm-related 9 Not determined
SPU_018859 Sp-Sarmr7 Sp-Sarm-related 7 Not determined
SPU_020008 Sp-Sarmr2 Sp-Sarm-related 2 Not determined
SPU_021841 Sp-Sarmr6 Sp-Sarm-related 6 Not determined
SPU_027639 Sp-Sarmr8_1 Sp-Sarm-related 8 Not determined
SPU_027640 Sp-Sarmr8 Sp-Sarm-related 8 Not determined
Novel family of TIR domain containing genes
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_003608 Sp-Tirc5 Sp-TIR-containing 5 Not determined
SPU_007952 Sp-Tirc2_1 Sp-TIR-containing 2 Not determined
SPU_012671 Sp-Tirc4 Sp-TIR-containing 4 Not determined
SPU_013299 Sp-Tirc2 Sp-TIR-containing 2 Not determined
SPU_013352 Sp-Tirc6(partial) Sp-TIR-containing 6 Not determined
SPU_014926 Sp-Tirc3 Sp-TIR-containing 3 Not determined
SPU_016014 Sp-Tirc1 Sp-TIR-containing 1 Not determined
SPU_020131 Sp-Tirc7 Sp-TIR-containing 7 Not determined
ECSIT/SITPEC
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_012096 Sp-Ecsit Sp-Sitpec Not determined
Tollip
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_026252 Sp-Tollip_1 - Not determined
SPU_021673 Sp-Tollip - Not determined
II.A3 Intracellular signal transduction - NLR signaling
PEPT1/slc15a1
Annotated Common Synonyms Ortholog/ Gene Names Homolog
SPU_012690 Sp-Slc15a2 Oligopeptide transporter, kidney isoform (Peptide transporter 2) (Kidney H(+)/peptide cotransporter) (Solute carrier family 15 member 2)
GRIM19/Ndufa13
Annotated Common Synonyms Ortholog/
Gene Names Homolog
SPU_003408 Sp-Ndufa13_1
SPU_024115 Sp-Ndufa13
NADH-ubiquinone oxidoreductase B16.6 subunit (Complex I-B16.6) (CI-B16.6) (Gene associated with retinoic-interferon-induced mortality 19 protein) (GRIM-19) (Cell death-regulatory protein GRIM-19)
NADH-ubiquinone oxidoreductase B16.6 subunit (Complex I-B16.6)(CIB16.6) (Gene associated with retinoic-interferon-induced mortality 19 protein)(GRIM-19) (Cell death-regulatory protein GRIM-19) -
RIPK1/RIP and RIPK2/RICK/CARDIAK
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_005215 Sp-Ripk1-4A - Not determined
CRADD- and ASC-like
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_011816 Sp-Ripk1-4B - Not determined
Caspase-1/4/5/11/12/13
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_019893 Sp-Pan1 Possible adaptor for NLRs Not determined
SPU_006410 Sp-Pan2 Possible adaptor for NLRs Not determined
SPU_002921 Sp-IceL1a - Not determined
SPU_002923 Sp-IceL1b - Not determined
SPU_011872 Sp-IceL3 - Not determined
SPU_012722 Sp-IceL4 - Not determined
SPU_021141 Sp-IceL2 - Not determined
II.B1 Intercellular signaling (Cytokines and Growth Factors) - Interleukins, Cytokines and Hematopoietins
MIF
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_001152 Sp-Mif7 - Not determined
SPU_011299 Sp-Mif1 - Not determined
SPU_012071 Sp-MifL1 - Not determined
SPU_016226 Sp-Mif6 - Not determined
SPU_017901 Sp-Mif3 - Not determined
SPU_019323 Sp-MifL2 - Not determined
SPU_020035 Sp-Mif4 - Not determined
SPU_020036 Sp-Mif5 - Not determined
SPU_030001 Sp-Mif2 - Not determined
IL-1R/CD121a and associated genes
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_003911 Sp-Il1ap - Not determined
SPU_003912 Sp-Il1ap_1 - Not determined
SPU_005871 Sp-Il1r1 IL1RA; CD121A; IL1R-alpha Not determined
SPU_013950 Sp-Il1r/Rs1 - Not determined
SPU_000409 Sp-Il1r/Rs1(d) - Not determined
Pellino
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_004517 Sp-Pellino - Not determined
Annotated Common Synonyms Ortholog/Homolog Gene Names
PU_005983 Sp-Il17-1 - Not determined
SPU_012844 Sp-Il17-2 - Not determined
SPU_012845 Sp-Il17-3 - Not determined
SPU_019349 Sp-Il17-4 - Not determined
SPU_019350 Sp-Il17-5
Not determined
SPU_019351 Sp-Il17-6 - Not determined
SPU_022838 Sp-Il17-7 - Not determined
SPU_027904 Sp-Il17-8 - Not determined
SPU_030141 Sp-IL17rL Sp-IL17R-like,Sp-IL17RA-like Not determined
SPU_030184 Sp-Il17-9
SPU_030185 Sp-Il17-10
SPU_030186 Sp-Il17-11
SPU_030187 Sp-Il17-12
SPU_030188 Sp-Il17-13
SPU_030189 Sp-Il17-p1
SPU_030190 Sp-Il17-14
SPU_030191 Sp-Il17-15
SPU_030192 Sp-Il17-16
Interleukin-17, Cytotoxic T-lymphocyte-associated antigen 8 Not determined
Interleukin-17, Cytotoxic T-lymphocyte-associated antigen 8 Not determined
Interleukin-17, Cytotoxic T-lymphocyte-associated antigen 8 Not determined
Interleukin-17, Cytotoxic T-lymphocyte-associated antigen 8 Not determined
Interleukin-17, Cytotoxic T-lymphocyte-associated antigen 8 Not determined
Interleukin-17, Cytotoxic T-lymphocyte-associated antigen 8 Not determined
Interleukin-17, Cytotoxic T-lymphocyte-associated antigen 8 Not determined
Interleukin-17, Cytotoxic T-lymphocyte-associated antigen 8 Not determined
Interleukin-17, Cytotoxic T-lymphocyte-associated antigen 8 Not determined
SPU_030193 Sp-Il17-17 Interleukin-17, Cytotoxic T-lymphocyte-associated antigen 8 Not determined
SPU_030194 Sp-Il17-p2 Interleukin-17, Cytotoxic T-lymphocyte-associated antigen 8 Not determined
SPU_030195 Sp-Il17-p3
SPU_030196 Sp-Il17-18
Interleukin-17, Cytotoxic T-lymphocyte-associated antigen 8 Not determined
Interleukin-17, Cytotoxic T-lymphocyte-associated antigen 8 Not determined
SPU_030197 Sp-Il17-19 Interleukin-17, Cytotoxic T-lymphocyte-associated antigen 8 Not determined
SPU_030198 Sp-Il17-20 Interleukin-17, Cytotoxic T-lymphocyte-associated antigen 8 Not determined
SPU_030199 Sp-Il17-21 Interleukin-17, Cytotoxic T-lymphocyte-associated antigen 8 Not determined
SPU_030200 Sp-Il17-22 Interleukin-17, Cytotoxic T-lymphocyte-associated antigen 8 Not determined
SPU_030201 Sp-Il17-p4 Interleukin-17, Cytotoxic T-lymphocyte-associated antigen 8 Not determined
SPU_030202 Sp-Il17-p5 Interleukin-17, Cytotoxic T-lymphocyte-associated antigen 8 Not determined
SPU_030203 Sp-Il17-23 Interleukin-17, Cytotoxic T-lymphocyte-associated antigen 8 Not determined
SPU_030204 Sp-Il17-24 Interleukin-17, Cytotoxic T-lymphocyte-associated antigen 8 Not determined
IL-17RA/B
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_030141 Sp-IL17rL Sp-IL17R-like,Sp-IL17RA-like Not determined
II.B2 Intercellular signaling (Cytokines and Growth Factors) - TNF pathway
TNF Superfamily
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_009527 Sp-TnfsfL2 tumor necrosis factor superfamily like2 Not determined
SPU_009528 Sp-TnfsfL1 tumor necrosis factor superfamily like1 Not determined
SPU_015654 Sp-TnfsfL3 tumor necrosis factor superfamily like3 Not determined
SPU_030072 Sp-TnfsfL4 tumor necrosis factor superfamily like4 Not determined
TNFR Superfamily
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_010180 Sp-Tnfrsf_cl1 tumor necrosis factor receptor superfamily classical1 Not determined
SPU_010230 Sp-TnfrsfL1 tumor nectosis factor receptor superfamily like1 Not determined
SPU_012211 Sp-Tnfrsf1a tumor necrosis factor receptor superfamily 1a, TNFR, TNFAR, TNFR1, p55-R, CD120a Not determined
SPU_018915 Sp-TnfrsfL5 tumor necrosis factor receptor superfamily like1 Not determined
SPU_020955 Sp-TnfrsfL2 tumor necrosis factor receptor superfamily like2 Not determined
SPU_026216 Sp-Tnfrsf_cl2 tumor necrosis factor receptor superfamily classical2 Not determined
SPU_020740 Sp-Eda2rL1 ectodysplasin A2 receptor, X-linked ectodysplasin receptor, tumor necrosis factor receptor superfamily member XEDAR, TNFRSF27 Not determined
SPU_024584 Sp-Eda2rL2 ectodysplasin A2 receptor, X-linked ectodysplasin receptor, tumor necrosis factor receptor superfamily member XEDAR, TNFRSF27 Not determined
FADD
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_010777 Sp-Fadd - Not determined
II.B3 Intercellular signaling (Cytokines and Growth Factors) - RTK signaling
Tie1/2
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_024044 Sp-Tie1/2 - Likely ortholog of Vertebrate Tie1 AND Tie2
Flk-1/Flt-1/Flt-4 (VEGFR1-3)
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_000310 Sp-Pdgfr/vegfrL - Urchin-specific homolog of gene family defined by GLEAN3_00310
SPU_021021 Sp-Pdgfr/vegfrL_1 - Urchin-specific homolog of gene family defined by GLEAN3_00310
VEGF
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_005737 Sp-Vegf2 vascular endothelial growth factor Likely ortholog of VEGF-C
SPU_014978 Sp-Vegf vascular endothelial growth factor Not determined
SPU_030148 Sp-Vegf3 - Not determined
II.B4 Intercellular
signaling (Cytokines and Growth Factors) - Cytoplasmic adaptors and signal transducers
IRAK
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_000073 Sp-Pik1 Sp-Pelle/Irak1 Not determined
SPU_028724 Sp-Pik2 Sp-Pelle/Irak2 Not determined
JAKs
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_022495 Sp-Jak1 Janus kinase 1 Not determined
SPU_030066 Sp-Jak_2 - Not determined
SOCS
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_002792 Sp-Socs2/3 - Unclear
SPU_010245 Sp-Socs7 - Likely ortholog of SOCS7 (Suppressor of Cytokine Signaling 7)
SPU_011298 Sp-Socs6L - Not determined
SPU_026496 Sp-Socs4/5 - Unclear
PIAS
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_007964 Sp-Pias1 GBP; DDXBP1; GU/RH-II Not determined
SPU_011690 Sp-Pias2/3 Protein inhibitor of activated STAT2 Not determined
Src Family Kinases
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_005419 Sp-Sfk3 Src Family Kinase, Yes-like kinase Likely ortholog of Asterina miniata SFK3
SPU_013522 Sp-Sfk1 Src, Src Family Kinase, Yes-like kinase Unclear
SPU_023261 Sp-Sfk1h Src, Src Family Kinase, Yes-like kinase Not determined
SPU_024525 Sp-Sfk7 Src Family Kinase, Yes-like kinase Not determined
SPU_026766 Sp-Sfk5 - Not determined
SPU_014473 Sp-Skf6 - Not determined
SPU_012805 Sp-Frk src-type protein tyrosine kinsase, SFK2, SFK4 Likely ortholog of HsFrk
Abl
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_023952 Sp-Abl v-Abl, c-Abl Likely ortholog of Asterinia miniata Abl
Syk/ZAP-70
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_006988 SpSyk/Zap70
WASP
Spleen tyrosine kinase, zeta-chain (TCR) associated protein kinase 70kDa, Srk
Likely ortholog of Syk (Spleen tyrosine kinase)
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_003194 Sp-Sim_wsk_ald_syn_ph WASP Not determined
SH2-B
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_027685 Sp-Sh2Brs1 - Not determined
SHP-1/2
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_013810 Sp-Ptpn6/11 SHP-2, PTP1D, PTP2C Not determined
FKBP-12
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_027840 Sp-Fkbp12 - Not determined
Nck
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_014752 Sp-Nck - Not determined
III. COAGULATION and WOUND REPAIR
III.a Coagulation cascade genes
CF5/8
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_021228 Sp-Cf5/8L1 Sp-Coagulation factor 5/8-like1 Not determined
SPU_021229 Sp-Cf5/8L2 Sp-Coagulation factor 5/8-like2 Not determined
CF13
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_005702 Sp-TransglutL - Likely ortholog of P lividus transglutaminase -like
TFPI
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_030138 Sp-TfpiL Sp-Tissue factor pathway inhibitor-like Not determined
Plasminogen
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_005414 Sp-PlasmgnL3 Sp-Plg-like3, Sp-Plg-l3 Not determined
SPU_014730 Sp-PlasmgnL2 Sp-Plg-like 2, Sp-Plg-l2 Not determined
SPU_014731 Sp-Unk_35 - Not determined
SPU_022934 Sp-PlasmgnL1 Sp-Plg-like1, Sp-Plg-l1 Not determined
SPU_000633 Sp-Plasmgnr1 Sp-Plg-related 1, Sp-Plg-r12 Not determined
Serpins (All Sp-serpin-like models Blast back to the B clade of mammalian serpins)
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_002711 Sp-SerpL11 serine (or cysteine) peptidase inhibitor, clade B (ovalbumin) Not determined
SPU_004543 Sp-SerpL10 serine (or cysteine) peptidase inhibitor, clade B (ovalbumin) Not determined
SPU_009346 Sp-SerpL2 serine (or cysteine) proteinase inhibitor, clade B (ovalbumin) Not determined
SPU_013377 Sp-SerpL3 serine (or cysteine) peptidase inhibitor, clade B (ovalbumin) Not determined
SPU_013378 Sp-SerpL4 serine (or cysteine) peptidase inhibitor, clade B (ovalbumin) Not determined
SPU_018196 Sp-SerpL8 serine (or cysteine) peptidase inhibitor, clade B (ovalbumin) Not determined
SPU_018630 Sp-SerpL7 serine (or cysteine) peptidase inhibitor, clade B (ovalbumin) Not determined
SPU_018631 Sp-SerpL6 serine (or cysteine) peptidase inhibitor, clade B (ovalbumin) Not determined
SPU_018632 Sp-SerpL5 serine (or cysteine) peptidase inhibitor, clade B (ovalbumin) Not determined
SPU_020278 Sp-SerpL12 serine (or cysteine) peptidase inhibitor, clade B (ovalbumin) Not determined
SPU_024263 Sp-SerpL9 serine (or cysteine) peptidase inhibitor, clade B (ovalbumin) Not determined
SPU_028469 Sp-SerpL1 PAI-2, Plasminogen Activator Inhibitor-2, PAI-2 Not determined
Plasma Kallikrein B1
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_012390 Sp-Klkb1 Plasma kallikrein precursor (Plasma prekallikrein) (Kininogenin)(Fletcher factor) [Contains: Plasma kallikrein heavy chain; Plasma kallikrein light chain]
Not determined
SPU_015878 Sp-Klkb1L1 Sp-Kallikrein B1-like1 Not determined
Alpha-2-Macroglobulin
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_011257 Sp-A2m Sp-alpha-2-macroglobulin Not determined
III.b Sea urchin clotting genes
Amassin
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_003389 Sp-OlfL Sp-amassin-like Unclear
SPU_006351 Sp-Olf Sp-amassin1 Not determined
SPU_021526 Sp-Ams amassin Not determined
SPU_023924 Sp-Olf_1 Sp-amassin2 Not determined
SPU_023926 Sp-Ams2 amassin2 Not determined
SPU_026884 Sp-Olf_2 Sp-amassin3 Not determined IV. MISCELLANEOUS
CD109
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_024565 Sp-Cd109L - Not determined
Serum amyloid protein
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_012667 Sp-Saa-a Sp-Serum amyloid A-a Not determined
SPU_012668 Sp-Saa-b Sp-Serum amyloid A-b Not determined
CD45
Annotated Common Synonyms
Gene Names
SPU_021599 Sp-Ptpr1 Sp-PTPR-G/Z
Unclear
Ortholog/Homolog
Immune related transcription factor gene models of the S. purpuratus genome
ETS
Ets-1,2
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_002874 Sp-Ets1/2 Ets1 Likely ortholog of human ets1 and ets2
Erg, Fli-1
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_018483 Sp-Erg Fli Urchin-specific homolog of gene family defined by GLEAN3_02874
PU.1, Spi-B, Spi-C
Annotated Common Synonyms
Gene Names
SPU_030060 Sp-Pu1 SpiB, SpiC
Elf-1
Annotated Common Synonyms
Gene Names
SPU_020123 Sp-ElfB E74
SPU_020124 Sp-ElfA E74
Tel
Ortholog/Homolog
Urchin-specific homolog of gene family defined by GLEAN3_02874
Ortholog/Homolog
Urchin-specific homolog of gene family defined by GLEAN3_02874
Urchin-specific homolog of gene family defined by GLEAN3_02874
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_008351 Sp-Tel_1 Yan Likely ortholog of ets variant gene 6
SPU_028479 Sp-Tel Yan Likely ortholog of ets variant gene 6
GABP
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_021557 Sp-Gabp - Urchin-specific homolog of gene family defined by GLEAN3_02874
GATA
GATA-1/2/3
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_027015 Sp-GataC - Likely ortholog of Gata-2/3
Basic Helix-Loop-Helix (HLH) - Class I bHLH
E2A, HEB, ITF-2
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_016343 Sp-E12 - Urchin-specific homolog of gene subfamily defined by transcription_factor_4_[Canis_familiaris]
Basic Helix-Loop-Helix (HLH) - Class II bHLH
SCL
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_028093 Sp-Scl - Urchin-specific homolog of gene subfamily defined by Tcell_acute_lymphocytic_leukemia_1_[Danio_rerio]
Basic Helix-Loop-Helix (HLH) - Class III bHLH
MITF/TFE3
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_008175 Sp-Mitf - Urchin-specific homolog of gene subfamily defined by transcription_factor_binding_to_IGHM_3a[D.rerio]
Basic Helix-Loop-Helix (HLH) - Class IV HLH/Id
Id-2
Annotated Common Synonyms
Gene Names
SPU_015374 Sp-Id - Not determined
Basic Helix-Loop-Helix (HLH) - Class VI Hairy/Hes
Hes-1,Hes-4
Annotated Common Synonyms
Gene Names
SPU_006813 Sp-Hairy Sp-HesB, hairy enhancer of split Unclear
Ortholog/Homolog
Ortholog/Homolog
SPU_006814 Sp-Hairy2/4 hairy, enhancer of split, e(spl), HES Not determined
SPU_015712 Sp-Hey4 hairy homolog
Likely ortholog of AAH12351.1| Hairy and enhancer of split 4 [Homo sapiens]
SPU_021608 Sp-HesC - Urchin-specific homolog of gene subfamily defined by hairy-related_6_[Danio_rerio]
Hey/HERP
Annotated Common Synonyms
Gene Names
Ortholog/Homolog
SPU_009465 Sp-Hey - Urchin-specific homolog of gene subfamily defined by hey1-prov_protein_[Xenopus_tropicalis]
Runx-1-3
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_006917 Sp-Runt1 SpRunt-1, SpRunt Urchin-specific homolog of gene family defined by Runt domain
SPU_007852 Sp-Runt2 SpRunt-2 Urchin-specific homolog of gene family defined by Runt domain
SPU_025612 Sp-Runt1_1 SpRunt-1, SpRunt Not determined
OLF/EBF
EBF-1,2,3
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_004702 Sp-Ebf3 EBF3, OE-2,Early B-cell factor 3 Likely ortholog of mouse EBF3
PAX
Pax-5
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_014539 Sp-Pax2-5-8 - Urchin-specific homolog of gene subfamily defined by paired_box_gene_2b_[Danio_rerio]
C/EBP
C/EBPalpha, beta, epsilon
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_001657 Sp-Cebpa CCAAT/enhancer binding protein alpha, C/EBP alpha
C/EBPgamma
Likely ortholog of CCAAT/enhancer binding protein alpha, C/EBP alpha
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_011002 Sp-Cebpg CCAAT/enhancer binding protein (C/EBP), gamma
Likely ortholog of CCAAT/enhancer binding protein (C/EBP), gamma
C2H2 Zinc Finger
Ikaros
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_011260 Sp-Z181 - Unclear
Egr-1,2,3
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_015358 Sp-Egr z60 Unclear
LKLF, EKLF
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_020311 Sp-Klf2/4 z85 Unclear Gfi-1
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_012645 Sp-Z166 - Unclear
Blimp-1
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_027235 Sp-Blimp1 Krox1a Not determined
TCF/LEF Hmg Box
TCF-1, LEF-1
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_003704 Sp-Lef1 Tcf
Urchin-specific homolog of gene subfamily defined by gene_is=HMG_protein_Tcf/Lef_[S.purpuratus]
SPU_027853 - - -
Interferon Response Factor
IRF-1,2
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_010404 Sp-Irf - Urchin-specific homolog of gene subfamily defined by interferon_regulatory_factor_1_[Gallus_gallus]
IRF-4/Pip, IRF-8/ICSBP
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_026877 Sp-Irf4 - Urchin-specific homolog of gene subfamily defined by interferon_regulatory_factor_4_[Gallus_gallus]
bZIP
c-fos, Fos, Fra-1, Fra-2
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_021172 Sp-Fra2 - Not determined
c-jun, JunB, JunD
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_003102 Sp-Jun - Urchin-specific homolog of gene subfamily defined by c-Jun_protein_[Danio_rerio]
ATF-2, ATF-7, CREB
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_026905 Sp-Atf2 - Urchin-specific homolog of gene subfamily defined by activating_transcription_factor_2_[Gallus_gallus]
XBP-1
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_008703 Sp-Xbp1 - Likely ortholog of X-box binding protein processed isoform [Mus musculus]
NFE
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_011174 Sp-Nfe2 - Urchin-specific homolog of gene subfamily defined by nuclear_factor_(erythroid-derived_2)-like_1[G.gallus]
REL Homology Domain c-rel, RelA, RelB
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_012203 Sp-Rel - Likely ortholog of C-rel [danio rerio]
NFkB1, NFkB p105
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_008177 Sp-Nfkb - Urchin-specific homolog of gene subfamily defined by gene_is=NFkB_protein_[Strongylocentrotus_purpuratus]
NFATc, NFAT-4
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_015908 Sp-Nfat - Likely ortholog of NFATz2 [mus musculus]
STATs
STAT5
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_015108 Sp-Stat signal transducer and activator of transcription Not determined
Nuclear Receptors
RARalpha
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_018366 Sp-Rara - Urchin-specific homolog of gene subfamily defined by CG18023-PA,_isoform_A_[Drosophila_melanogaster]
RARbeta
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_013178 Sp-Nr1m3 - Urchin-specific homolog of gene subfamily defined by retinoic_acid_receptor,_beta_[Gallus_gallus]
SOX Hmg Box
SOX4, SOX11, SOX22, SOX24
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_002603 Sp-SoxC Sox4/11/22/24 Urchin-specific homolog of gene subfamily defined by sox11_[Xenopus_tropicalis]
T-BOX
T-bet
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_025584 Sp-Tbr - Likely ortholog of Tbr1/Tbx21/Eomes
FORKHEAD
FoxP1,2,3,4
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_009876 Sp-FoxP - Likely ortholog of FoxP
SMAD
SMAD4
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_004287 Sp-Smad4 - Urchin-specific homolog of gene subfamily defined by MAD_homolog_4_isoform_1_[Mus_musculus]
SMAD2,3
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_017642 Sp-Smad2/3 - Urchin-specific homolog of gene subfamily defined by MAD_homolog_3_[Danio_rerio]
SMAD1,5
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_020722 Sp-Smad1/5/8_1 - Urchin-specific homolog of gene subfamily defined by MAD_homolog_1_[Xenopus_tropicalis]
SPU_023107 Sp-Smad1/5/8 - Urchin-specific homolog of gene subfamily defined by SMAD,_mothers_against_DPP_homolog_5_[Gallus_gallus]
DEAD RINGER/BRIGHT
Dead ringer
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_005718 Sp-Dri_2 - Likely ortholog of Human DriL1, DriL2
SPU_025601 Sp-Dr_1 - Likely ortholog of Human DriL1, DriL2
POU Domain
Oct1, Oct 2
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_009262 Sp-Oct1-2 Pou2F
Urchin-specific homolog of gene subfamily defined by major_octamer-binding_protein_[S.purp]
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_000861 Sp-Myb SpMyb, Myb-like transcription factor
Transcriptional Co-factors
LMO1/2
Urchin-specific homolog of gene subfamily defined by Myb DNA binding domain
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_013569 Sp-Lmo2t - Not determined
FOG1/2
Annotated Common Synonyms Ortholog/Homolog
Gene Names
SPU_010168 Sp-Fog - Likely ortholog of FOG1





