International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056

Volume: 09 Issue: 09 | Sep 2022 www.irjet.net p-ISSN: 2395-0072

International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056

Volume: 09 Issue: 09 | Sep 2022 www.irjet.net p-ISSN: 2395-0072
1,2,3,4Final Year, Dept. of Computer Engineering, R.M.D Sinhgad School of Engineering, Maharashtra, India 5H.O.D, Dept. of Computer Engineering, R.M.D Sinhgad School of Engineering, Maharashtra, India ***
Abstract - Brain tumor detection is one of the most complex biomedical problems. Its anatomical structure makes it complex to cure neuro medical issues. Medical segmentationis a challenging part in curing intense brain tumors. In such scenarios, deep learning algorithms are used to resolve the complexity of segmentation and detect the tumor accurately. The Convolutional Neural Networks (CNN) has been developed by the efficient auto segmentation technology. UNET is used along with methods of computer vision to increase the rate of successfully detecting the tumor. Use of biomedical image segmentation extensively has resulted in high rates of curing the tumor accurately. In this paper, we are proposing a multimodal brain tumor segmentation using 3D UNET. We have used the BraTS 2020 dataset which contains 369 3D MRI images that are used for training while 125 MRI images that are used for testing. We have developed a 3D model which generates the output in 3D format and were able to achieve accuracy of 98.5%.
Key Words: BrainTumor,BraTS2020,CNN,Gliomas,MRI, UNET,Segmentation.
Braincancertreatmentrequireshighexpertiseandprecision asitisoneofthemostcomplexproceduresinthebiomedical field. Deep learning is a class of machine learning. Deep learning algorithms use multiple layers in order to solve complexproblems.Inthispaper,wehaveusedmethodslike CNN:UNETtoperformsegmentationoverthetumoraffected area of the brain. The main problem of detecting a brain tumorisitsareaofinfectionaswellastheintensity.Hence, braintumorsegmentationisdone.Forimagesegmentation, themethodofimageinpaintingisused.Imageinpaintingisa methoddonepriortoUNETinordertoenhancetheimage quality. It helps to smoothen the image. This helps in identifying the tumor more accurately with the help of similarity index. Using the 3D MRI images given in the dataset,wehaveimportedthemtothesystemto perform UNET and perform segmentation to acquire the accurate tumor present in the brain. Mathematical models such as Edge-Based Detection Method, Rough Set-Based Fuzzy ClusteringandCross-EntropyLossFunction.Withthehelpof thesemodels,segmentationisdoneprecisely.
Various researches clearly depict the importance of deep learning methods for learning and identifying specific
patternsforaccuratecharacterizationofaspecificproblem. IntheRecentstudyperformedbyLingTanandWenjieMa[1] withinthecontextofmedicalimagesegmentationintheyear 2021theyhaveproposedamultimodalbraintumorimage segmentation method based on ACU-Net network using differentvolumesofdatasetsuchasBraTS2015,BraTS2018, BraTS2019.Thisstudydefinitelyshowedanincreaseddice, recall,precisionandcanemployanactivecontourmodelto overcomeimagenoiseandedgeslitstobetteridentifylowcontrastandlow-resolutionin the image but This studyis alsopresentedwithpooradaptability.
Asweknowthatgliomasaremalignantandheterogeneous Mahnoor Ali,Asim Waris proposed an ensemble of two segmentationnetworks3DCNNandaU-net[2]whichhelps us for better and accurate predictions. In this study researchersusedBraTS2019challengedatasetandachieved a good dice score of 0.91 for whole tumor region and provided efficient and robust tumor segmentation across multiple regions. There were certain discrepancies and deficiency as the recognition rate was only 83.4% hence betterpreandpostprocessingofdatawasrequired.
The most formal and important step for brain tumor segmentationistodetecttheboundaryofthetumori.e.,edge detection.Agroupof researchers namedAhmed H.AbdelGawad,LobnaA.SaidproposedaSystemforOptimizededge detectionincaseofbraintumordetectionbyanalyzingMRI images[3].ItusesdifferentTechniquesandAlgorithmssuch asBalance contrastEnhancement Technique(BCET),skull stripping and Genetic Algorithm (GA) which comes under evolutionarysciences.
Forproperdiagnosisandplanningforbraincancertreatment properbraintumorsegmentationisveryimportantsointhe year2020certainresearchersnamedNagwaM.Aboelenein, Piao Songhao, Alam Noor proposed an architecture called HybridTwoTrackU-net(HTTU)whichisusedforAutomatic BrainTumorSegmentation[4].Itusesdifferentmethodssuch as N4ITK Bias Correction, Focal loss and Generalized dice scorefunctions.ItusesBraTS2018datasetandalsoachieved 0.865dicescoreforwholetumorbutitcouldnotidentifythe underlyinglayersoftheimagesused.
Byusingfeaturefusiononseverallevelswecanmakefulluse of hierarchical features to reveal the importance of refinement and aggregation of features in brain tumor segmentation.Dongyuan Wu,Yi Ding,Mingfeng Zeng [5] developed a Multi Features Refinement and Aggregation
International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056

Volume: 09 Issue: 09 | Sep 2022 www.irjet.net p-ISSN: 2395-0072
(MRA)Modelforimprovementofsegmentationaccuracy.It uses BraTS 2015 dataset and achieves 0.83 dice score for wholetumorbuthaspoorperformanceincaseofenhancing tumor.
YiboHanandZhengZhangdevelopedanImageInteractive framework for Brain Image Segmentation [6] which is assisted by Deep Learning concepts. It uses Deep Convolutional Neural Networks (DCNN) which is a MultiTask Deep Learning Approach (MTDLA) but it produces minormistakesduetounsupervisedfinetuning.
DuringBrainTumorSegmentationbasedonMRIModalities certain noisy regions affect the accuracy and performance greatly. Guohua Cheng and Hongli Ji developed an system whichisusedtoevaluateAdversarialPerturbationbasedon MRIModalities[7]butwhenfourmodalitiesareattackedor damaged we will observe a severe degradation of performanceandaccuracywilloccur.
Toimprovetheperformanceofmedicalimagesegmentation andenhancementoflocalfeatureextractionJianxinZhang, ZongkangdevelopedanAttentionGateResU-netmodelfor Automatic Brain Tumor Segmentation [8]. They used different dataset volumes such as BraTS 2017,2018 and 2019.ItsurelyoutperformbaselinesofU-netandResU-net but loses an amount of context and local details among differentslices.
For proper extraction of Image features in brain tumor segmentation Weiguang Wang,Fanlong Bu developed an explanatorymodelwhichcombinesLearningMethodsofCNN withfeatureextractionofImagesinBrainTumordetection [9]. It is used for performance analysis, diagnostic reports and as theoretical references for any related research. DatasetusedisGBMdatavolumesanditachieves0.872dice scoreforwholetumorbuthashighcomputationaltimeand complexity.
ForbetterSupervisionandExcitationfunctionPingLiu,Qi Dou, Qiong Wang developed an Encoder-Decoder Neural Network[10]containing3DSqueezeandExcitationandalso includingDeepSupervisionforBrainTumorSegmentation. They used BraTS 2017 dataset and N4BiasFieldCorrection algorithmalongwithV-net(DSSE-V-net).Certaindrawbacks were found such as limited size kernel and receptive field problem.
There are various types of neural networks which can be used for Brain Tumor Segmentation, Wu Deng, Qinke Shi proposed a Deep Learning based model for brain tumor segmentation which uses two neural networks HeterogeneousConvolutionalNeuralNetworks(H-CNN)and CRF-RecurrentRegressionbasedNeuralNetwork(RNN)[11]. ItusestheBraTS2017datasetforevaluationandachieves highaccuracyandrecognitionrate.
TamjidImtiaz,ShahriarRifatdevelopedanAutomatedBrain TumorSegmentationsystemwhichisbasedonMulti-Planar SuperpixelLevelFeatureExtraction[12].Toreducethebias inintensitiesanIntensityAdjustmentSchemeisappliedon thewhole3DMRIimages.ExtremelyRandomizedTreesare usedforclassificationoftumorandnon-tumorclasses.This studyusesNCI-MICCAI2013challengedatasetwithcertain drawbacksfoundsuchaslowlevelofprecisioninthetumor regionsegmentation.
As most medical imaging dataset whichare used for brain tumor segmentation are small and fragmented Changhee Han,LeonardoRundo,RhyosukeArakidevelopedasystemas Brain Tumor Augmentation for Tumor detection which is achievedbycombiningNoise-to-ImageandImage-to-Image GAN[13].ItusesBraTS2016datasetwithResNet-50andtSNEmethodsbutresultsinpooroptimizationresults.
Theclassificationofbraincancerbymanualmethodrequires the knowledge and expertise of a Physician who is experienced enough in the medical field. Abdu Gumaei, MohammadMehediHassancameupwithaHybridFeature Extraction Method for Brain Tumor Segmentation that includes implementation of Regularized Extreme Learning Machine(RELM)[14].BraTS2013dataset,FuzzyC-means algorithm and Cellular Automata with a Grey Level Cooccurrence matrix (GLCM) are used in this study and relativelylowefficiencyispresent.
Cascading is a very popular concept in neural networks classes.KaiHu,QinghaiGan,YuanZhangdevelopedasystem for brain tumor segmentation which uses Multi- Cascaded ConvolutionalNeuralNetworks(MCCNN)andConditional Random Field (CRF) for its implementation [15]. When integrating into 3D CNN Poor effectiveness of images is observed.
Outofthemultipleimagingtechniquesusedtodetectbrain tumors MRI is commonly used as it has superior image quality with highest resolutions are obtained without any radiation. In the year 2019, Hossam H. Sultan, Nancy M. Salem,WalidAL-AtabanyproposedaDeepLearningmodel which is based on multi-classification method for Brain Tumor Images [16]. It uses Nanfang Hospital and General Hospital and The Cancer Imaging Archive (TCIA) public accessrepositoryforevaluationoftheproposedmodel.
AiminYang,XioleiYang,WenruiWudidaresearchstudyin theyear2019basedonFeatureExtractionofTumorImage using Convolutional Neural Networks (CNN) [17] which concludedthatCNNalgorithmshowshighaccuracyintumor image feature extraction and also demonstrated different advantagesofCNNinneuroimagingfield.Inthisstudylocal binary model algorithm andconvolutional neural network algorithmareusedtoextracttherequiredfeaturesoftumor.
International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056

InrecentyearstheapplicationsofAIinMagneticResonance Imaging (MRI) have been applied in various biomedical studies. In the year 2018 Gunasekaran Manogaran, P. Mohamed Shakeel developed a Machine Learning Based Approach for Brain Tumor Detection and data sample imbalanceanalysis[18].Itcontainsanimprovedorthogonal gammadistribution-basedmethodwhichisusedtoanalyze the under-segments and over-segments of brain tumor regionstoautomaticallydetectabnormalitiesintheRegionof Interest(ROI)butithaslowreal-timeapplications
Intheneurosciencefield,differentAlgorithmsareusedfor preciseandaccuratebraintumorSegmentation.QingnengLi, Zhifan Gao proposed a Unified Algorithm for Glioma SegmentationusingMultimodal MRIimages[19]. Itusesa TwoStepRefinementStrategytomaintainPPVvalueswhich inturnincreasesprocessingtimeandreducestherecognition rate.
Guotai Wang, Wenqi Li, Maria A. Zuluaga developed an InteractiveMedicalImageSegmentationmodelwiththehelp of Image-Specific Fine Tuning [20]. This model uses PretrainedGaussianMixtureModel(GMM)soithasfeweruser interactionsandless user time thantraditional interactive segmentation methods. But this model requires a large numberofannotatedimagesfortraining.
CNN(ConvolutionalNeuralNetwork),BraTS(BrainTumor Segmentation), MRI (Magnetic Resonance Imaging), CRF (ConditionalRandomFields),GAN(GenerativeAdversarial Networks),PPV(PositivePredictiveValue).
Thesurveydiscussedhereaimsfortheliveimpactofother methodologiesinBrainTumorsegmentationearlier.Inthis survey,hospitalswhicharecurrentlyworkingondetection andsegmentationofBrain Tumor basedon this particular methodgivesusaclearideaabouttheprocedureandoverall desiredresultsacquiredduringtheirprocess
Inordertoidentifythesub-regionssuchasEdema,enhancing tumor,andNecrosisthisTataMemorialHospitalproposeda 3D UNET architecture which helped in identifying these radiological sub-regions of a Brain Tumor. To keep the balancebetweenthetumorandnon-tumorpatchesinsidea brain,patchextractionschemehasalsobeenputforwardto addresstheproblem.Andalongwithwhichthearchitecture helps for precise location of a Tumor by using symmetric expandingfeaturesandtocapturethecontextbycontracting path. The desired results achieved during the process contains the Dice Scores of 0.92, 0.90 and 0.81 for Whole Tumor(WT),TumorCore(TC)andEnhancingTumor(ET), respectivelybasedonindependentpatients’datasetfromthe hospital.
PresentworkinApolloHospitalgivesinsightaboutavisual and quantitative analysis on automated Brain Tumor segmentation using Fuzzy-Possibilistic C-Means (FPCM) methodology. The K-means w.r.t patch-based technique is alsousedtocarryouttheskullscripting,alongwithRegionof Interest(ROI) it is possibleto measure the quantityof the exactregionofabraintumorlocated.Theresultsinvolved are improved based on performance achieved than the previousalgorithmsusedtocarryoutthesamebenchmark results.
The dataset used for our system is BraTS2020 Dataset (Training+Validation).ItcontainsMulti-MagneticResonance Imaging (MRI) scans. The Dataset mainly focuses on the segmentation of an essential heterogeneous brain tumor, namely gliomas. It is also used to evaluate the uncertainty betweendifferentalgorithmsintumorsegmentation. DatasetReference-www.kaggle.com
Size-40GB
No.ofTrainingSets-369
No.ofValidationSets–125
Beforeloadingthedatasetintothetrainingmodel,wehaveto perform data pre-processing operations to remove noisy regions and extract key characteristics required for the segmentation. BraTS 2020 does not contain any missing valuesorsetofMRIimagesforanypatientandallthedata fields are well labelled. The dataset is split into two parts: TrainingandTestingforfurtherevaluation.
With the survey done above there are certain remarkable discrepancies and inefficiencies with regards to the segmentationaccuracywhichfoundtobelowduetolackof efficientalgorithms.Pre-processingofthedata isnotdone due to which the dataset might contain noisy regions and erroneous values in-turn affecting the accuracy and recognition rate. Along with which boundary and depth detectionoftumorsisnotaccuratewhichmakesitdifficultto perform necessary actions. Also, the process is semiautomated with a low performance index that involves manualwork.Thus,poorresultsobtainedwithregardstothe enhancing tumor region. Our proposed system overcomes certainproblemsidentifiedinthepreviouswork.Withthis proposed system, we aim to increase the accuracy of the BrainTumorsegmentationbyusingabettertrainingsetand algorithmicpatternsfortheWholeCore(WC)andEnhancing
Volume: 09 Issue: 09 | Sep 2022 www.irjet.net p-ISSN: 2395-0072 © 2022, IRJET | Impact Factor value: 7.529 | ISO 9001:2008 Certified Journal | Page591
International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056

Volume: 09 Issue: 09 | Sep 2022 www.irjet.net p-ISSN: 2395-0072
tumorregion.ProjectingtheTumorregionissegmentedin 3D format for better understanding of underlying layers, depth,location,sizeandlength.Also, better preprocessing and post-processing of data has been done for efficient output.
After the completion of Requirement gathering and RequirementAnalysis,wearriveatthedesignstageofour implementation. The System Architecture represents the overall functionalities, modules, process flow, and a brief overview of the complete system through the eyes of a developer. Different modules have different assigned functionalitieswhichareimplementedinadefinedmanner with supervised control flow. In our proposed system we have converted the tumor detection operation into small modules which increases modularity and also strikes out variousinterdependenciespresentincaseofsimultaneous execution. Fig. 1 shows the System architecture of our System.
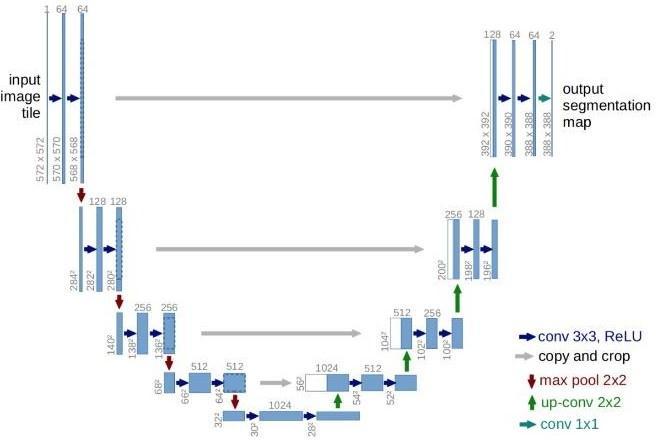
U-Netisanadvancedversionofasophisticatedconvolutional neural network (CNN) that was specially developed for biomedical image segmentation. The feature about U-Net whichdiffersfromCNNisthatthetargetisnotonlytoclassify whetherthereisaninfectionornotbutalsotoidentifythe areaofinfection.U-Netyieldsmoreprecisesegmentationby using comparatively fewer high-resolution images than previously used FCN (Fully Convolutional Networks) and othersegmentationmodels.Italsodoesnotrequiremultiple runsandalabelledsetofimagesforsegmentation.U-Netis divided into three main layers namely, Contraction Path, BottleneckandExpansionPath.Inordertowork onfewer parameters Max Pooling operation is carried out which reduces the size of the feature map. In this process, the popularactivationfunctionReLU(RectifiedLinearActivation Function) has been used which outputs the positive input directlybutoutputszeroiftheinputisnegative.
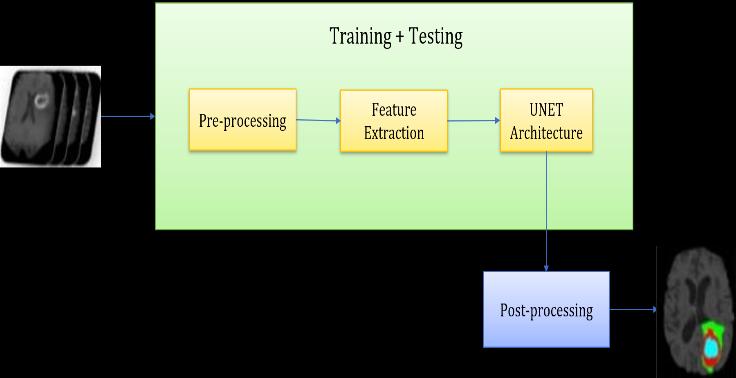
Thedatasetusedcontainsasetof3DMRIimagesofevery individual patient which are used as input for our system. Themainfunctionalblockisnamedas“Training+Testing” which contains different operational modules which are interlinked and follows a sequential pattern for its implementation. First sub-module is used for Data Preprocessing which includes operations such as Data cleaning,DataTransformationetc.Thenextmodulefollowed by Data preprocessing is Feature Extraction, it contains different operations such as skull stripping, boundary detection algorithms, cluster analysis, intensity, slicing methods etc. which are used for accurate delineation of Gliomas.Afterthecompletionofthefirsttwosub-processes, we provide the outcome as input images to the U- net Architecturewhichcontains a definedset ofconvolutional layer information and algorithmic conditions specified for segmentation.Theoutcomereceivedafterexecutionofthe “Training+Testing”functionalblockweprovidethisasan input for Data Post-processing which provides us only the segmentedportionofbraintumorasanoutput.
Fig. 2 –UNETArchitecture
For implementation of any algorithmic model certain mathematicalconstraintsmustbesatisfiedtoensureproper structure and provide strong calculative measures for productiveassessmentandevaluationofoutcomesreceived.
Edgebaseddetectionitselfincludesmultiplemathematical methodsthatidentifythecoordinatepixelsofanMRIscanat whichtheimagebrightnessmightshowsomediscontinuities. The pixels at which the brightness changes sharply are clusteredintoasetofcurvedlinesegmentscallededges.In case of Brain Tumor Segmentation, we can drastically improvetheperformanceofanyneuralnetworkappliedifwe firstdetecttheboundaryofthewholetumorregioninsteadof applyingthenetworkonacompleteMRIimage.Ifweapply U-netarchitectureonacompletebrainMRIimagethenthe processing time will be considerably high, the recognition
International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056

Volume: 09 Issue: 09 | Sep 2022 www.irjet.net p-ISSN: 2395-0072
Inourproposedsystem,wehavesuccessfullyextractedthe segmentedbraintumorimages.Weadoptedthemethodof UNETtoextractthesegmentedtumorintheformatoftwodimensional(2D)aswellasthree-dimensional(3D).Thebase architectureusedisthesameforbothmethods2D&3D.In this section working functionalities, evaluation metrics, performance scores, graphical representation of trends in caseofparametersusedareshowninadetailedmanner.To measurethesegmentationeffectofUNET,thispaperadopts subjectivevisualevaluationaswellasmanualcalculationsto judge the experimental results. The results section will be divided in two parts for precise articulation of outputs achievedforoursystem
A rough set-based fuzzy clustering consists of two steps, initialclusteringbasedonroughsetandsecondaryclustering basedonfuzzyequivalencerelations.TheRSFCLalgorithm haspreferableclusteringvalidityandhighrunefficiencyin handlingtheclusteringproblemsofbothnumericaldataand nominal data. In case of Brain Tumor Segmentation after successfully detecting the tumor boundary by using edgebased detection method, we use rough set to locate pixels whichareincludedintheisolatedregionformedafterEdge detectionandFuzzyClusteringtoformclustersofpixelson thebasisofsimilarityinintensityvariation.Thisclusteredset ofpixelsalsoresidesintheregionconfinedinsidethetumor boundary. …Eq. 4
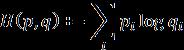
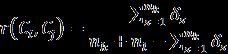
Crossentropylossisalsocalledlogarithmiclossorlogloss,it is usedalong withU-netArchitecturetominimize the loss occurred during training. Cross entropy is the most commonly used loss function in machine learning as it measures the performance of a classification model by consideringoutputasaprobabilityvaluebetween0and1
6.1.1 2D EVALUATION
6.1.1.1
InoursystemwehaveusedPython3.7astheprogramming language throughout the modules as it provides a vast support of libraries and also increases productivity. For training the deep learning model we have used keras as a backend with TensorFlow support. Keras contain various layers which are basic building blocks of neural networks. From layers certain functionalities are imported such as BatchNormalization,ActivationfunctionandConvolutionetc. Adam optimizer is used for the training model as it is the mostefficientoptimizerincaseofconvolutionaryoperations. Forfittingthetrainingmodel,thebatchsizeiskeptas32and epocharesetto40.Afterthecompletionoftraining,wesave the weights of the trained model in HDF5 (.h5) format for furtherevaluation
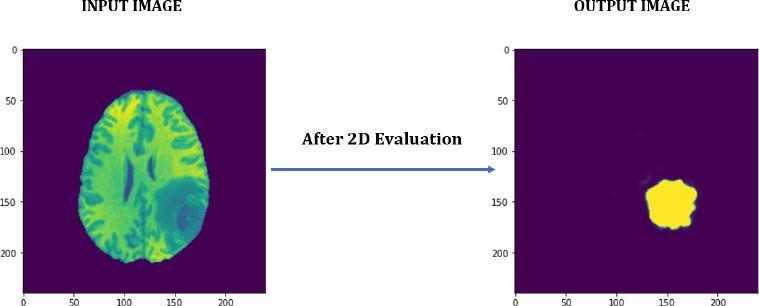
With the proposed architecture, the segmented tumor is extractedintheformof2Dimages.Fig.3showstheimageof tumorextracted.
…Eq. 5
…Eq. 6
Fig. 3 –2DOutput
RecognitionRate(Sensitivity/Recall):Itisdefined asameasureonhowwellasystemcanidentifytrue
International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056

Volume: 09 Issue: 09 | Sep 2022 www.irjet.net p-ISSN: 2395-0072
positivevalues.ItisalsonamedasTRUEPOSITIVE VALUE.Oursystemhasachievedarecognitionrate of96.13%. …Eq. 7
Specificity: Specificityisthemeasureoffindingthe proportion of actual negative values which are predicted negative. Hence, it is also called TRUE NEGATIVEVALUE.Thespecificityproducedforthe proposedsystemis99.91% …Eq. 8
Precision: PrecisionistheratiobetweentheTrue positiveandall thepositives.Amountofprecision recordedis92.30%. …Eq. 9
Accuracy: Accuracy is defined as the number of correctlypredictedvaluesfromallthedatavalues. The overall accuracy for the proposed system is measuredas99.10%.
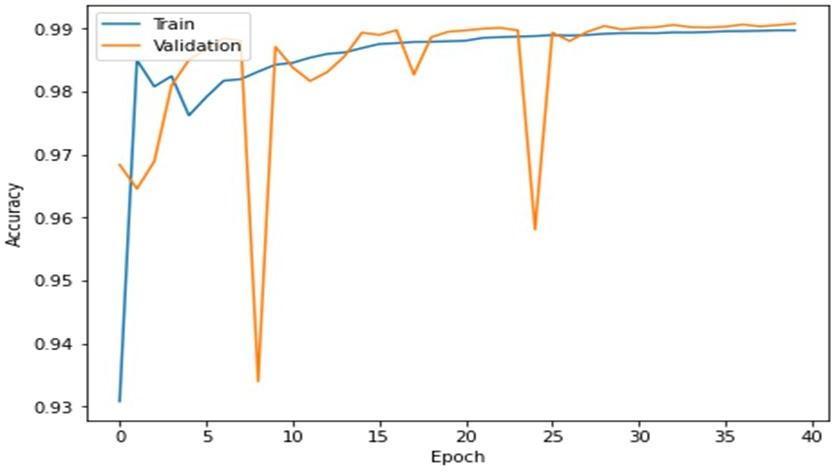
Fig.4showstheconstantandminimalincreaseinaccuracy overeverysubsequentepochcycle.
certainprerequisitesthatwehavetofulfillbeforejumping intothecodingstage.Theseprerequisitescanbecategorized as software requirements and hardware requirements depending upon the platform used for implementation. In caseofDeeplearningmodels,astrongprocessorcapableof complexcomputationisrequiredtoreducetheprocessing timeandincreasetheefficiencyalongwithaccuracy.Inthe general scenario, GPUs with CUDA enabled processors are usedfortrainingofneuralnetworksandmachinelearning models.CertaincomputationscanbeperformedonCPUwith moderate RAM before training the model. The hyperparameters can be manipulated for various runs to capturedifferentbehaviorofthemodelastrainingthemodel toachievehighaccuracyandprecisionmayrequiretrialand runagainapproachbefore,weoverfitorunderfitthemodel. Whenitcomestosoftwarerequirementsandspecification the compatibility and supportability of programming language along with IDE used is very important. Also, if someonedesirestotrainaparticularneuralmodelonGPU forbetterperformancethenCUDAcompatibilityandsupport for present NVIDIA drivers must be checked and verified beforegettingintoanyfurtherprocesses.Nowadaysthereare various options to integrate our system on a cloud-based architecturewhichdiminisheshighcomputationalhardware requirementscompletely.
In 3D evaluation, we have used Python 3.6.3 language for programming for individual components. Also, for neural networks we have used PyTorch, Keras backend with TensorFlowsupportetc.WehavecreatedasimpleGUIfor betteruserinteraction,inwhichtheuserisabletonavigate tabssuchasBrowsingtheInputImagefileofanyindividual fromthedatasetandsegmentittoobtainthetumorresult, accordingly, the segmented tumor in 2D can be converted intoa3Dstructurewiththehelpoftheothertabmentioned7 below and finally, by clicking the last tab that says “Show Result” gives the output ofa 3D Tumor whichcan be seen fromaBird’s-Eyeview.
After successfully evaluating the 3D model the output obtained from input 3D MRI are also represented in 3D format.Basedontheintermediatesnapshotsgeneratedfor differentplanesaftertheevaluationofthemodel,themost stable plane isselectedto display thetumorsegmentation whichisshowninFig.5.
Fig. 4 –Accuracyvs.Epoch
Forimplementationofthemodelproposedwhichgenerates theoutputresultofsegmentedtumorin3Dformatthereare
International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056

Volume: 09 Issue: 09 | Sep 2022 www.irjet.net p-ISSN: 2395-0072
TorepresentthegeneratedtumorsegmentationFig.5,in3D format we convert the stored information about gradual planesanddisplayitinBird’s-Eyeviewform.Fig.7clearly depictsthedesiredoutputofanisolatedtumorregionin3D. The3Doutputgeneratedcanbestoredonthelocaldevicein GIF or MP4 format to further utilize it for any biomedical studybasedonbraintumorsegmentation.
Aftersuccessfullytrainingthemodelcertainparametersare generated which are also known as performance scores. Some of the parameters with high significance are Dice coefficient (DSC), Recognition Rate (Sensitivity/Recall), SpecificityandAccuracy.
DSC also known as F1 Score is 0.579 along with the RecognitionRateof0.9106andSpecificityas0.9857forour proposed system. After the complete evaluation the ACC attainedis98.48% …Eq 10 …Eq. 11 …Eq 12 …Eq 13
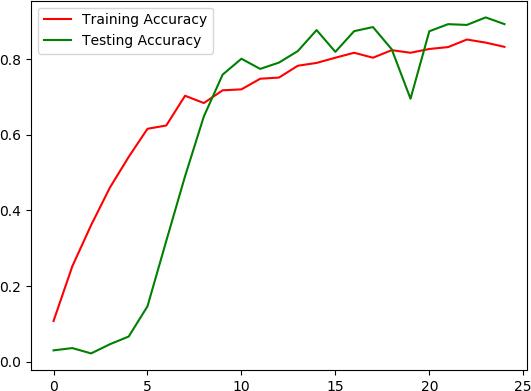
TheFig.6givesusvisualrepresentationofAccuracyvarying oversubsequentepochs.
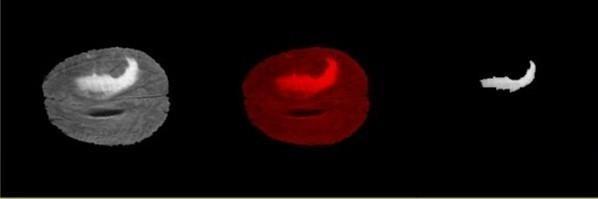

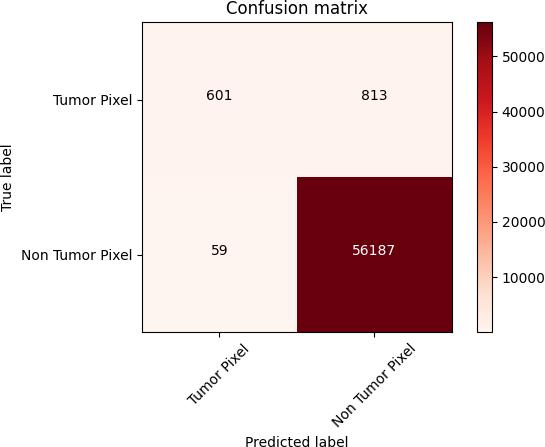
AConfusionmatrixwhichisalsoknownaserrormatrixisa table that is often used to describe the performance of a classificationmodel,here,theclassificationisbetweentumor pixelsandnon-tumorpixelsclasses.Confusionmatrix8isa special type of contingency table that contains two dimensions actual and predicted values which gives an identicalsetofclassesknownasTP(TruePositive),FP(False Positive),TN(TrueNegative)andFN(FalseNegative).These values summarize the results of our classification and can alsobeusedtoderiveotherperformanceparameterseither throughmanualcalculationorfetchingtheresultstoanother model.
International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056

Fig. 8 –ConfusionMatrix
Fig. 8 represents the values of identical set of classes generatedforoursystem.
TP = 601 FP = 813
FN = 59 TN = 56187
Using these values, we have calculated other performance parameters such as NPV (Negative Predictive Value), FNR (FalseNegativeRate),FPR(FalsePositiveRate),FDR(False DiscoveryRate),FOR(FalseOmissionrate),MCC(Matthews CorrelationCoefficient).
In this paper, we have successfully implemented U-Net Architecture for precise and accurate brain tumor segmentation.Thealgorithmtakesvarioustrainingimages provided in BraTS 2020 dataset and uses its predefined convolutional patterns to determine the tumor region effectively. Different mathematical models and strategies suchasEdgebaseddetectionmethod(EBD),Roughset-based Fuzzy Clustering, Cross Entropy Loss are applied in an abstractwayforcalculatedprocessflowandoperations.We aimtoaccuratelyextractkeyfeaturesandcharacteristicsfor thebraintumorbyproposingthismultimodalbraintumor detection and segmentation system. By this system, we developedamodeltorepresentthewholeoverviewofthe brain tumor region. Experimental results prove that our systemhasahighACCof99.10%incaseof2Devaluationand 98.48%for3Devaluation.
Withregardstothefuturescopeofourproposedsystem,we aimtoobtaintheseverityofBrainTumoralongwiththetype ofTumor.RecognitionofSurvivalRateswilladdvaluetothe system as well as for immediate treatment of a patient. Detectingthegrowthpatternwillhelptocurbthespreadof BrainTumor.
[1] W.M.J.X.LINGTAN,"MultimodalMagneticResonance Image Brain Tumor Segmentation Based on ACU-Net Network,"IEEE,vol.9,pp.14608-14618,Jan2021.
…Eq 14
…Eq 15
[2] S. O. G. A. W. Mahnoor Ali, "Brain Tumor Image Segmentation Using Deep Networks," IEEE, vol. 8, pp. 153589-153598,Aug2020.
[3] A. h. Abdel-Gawad, "Optimized Edge Detection Technique for Brain Tumor Detection in MR Images," IEEE,vol.8,pp.136243-136259,Jul2020.
…Eq 16
…Eq 17
[4] P. S. Nagwa M. Abouelenein, "HTT UNET: Hybrid Two TrackU-Netforautomaticbraintumorsegmentation.," IEEE,vol.8,pp.101406-101415,May2020.
[5] Y.D.M.Z.DongyuanWu,"MultiFeaturesRefinementand AggregationforMedicalBrainSegmentation,"IEEE,vol. 8,pp.57483-57496,Mar2020.
…Eq 18
Asaresult,theperformanceparametersNPV,FNR,FPR,FDR, FOR,MCCobtainedwiththehelpofsetofclassesgenerated for the systemare 0.9989, 0.0893, 0.0142, 0.5749, 0.0010, 0.6164respectively.
[6] Y. H. a. Z. Zhang, "Deep learning assisted image interactiveframeworkforbrainimagesegmentation," IEEE,vol.8,Jun2020.
[7] H. Guohua Cheng, "Adversarial Perturbation on MRI ModalitiesinBrainTumorSegmentation,"IEEE,vol.8, pp.206009-206015,Oct2020.
Volume: 09 Issue: 09 | Sep 2022 www.irjet.net p-ISSN: 2395-0072 © 2022, IRJET | Impact Factor value: 7.529 | ISO 9001:2008 Certified Journal | Page596
International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056

[8] J. D. Jianxin Zhang, "Attention Gate Res U-Net for AutomaticMRIBrainTumorSegmentation,"IEEE,vol.8, pp.58533-58545,Mar2020.
[9] F. B. ,. Z. L. WEIGUANG WANG, "Learning Methods of Convolutional Neural Network Combined With Image FeatureExtractioninBrainTumorDetection.,"IEEE,vol. 8,pp.152659-152668,Aug2020.
[10] Q. D. Q. W. PING LIU, "An EncoderDecoder Neural Network With 3D Squeeze-and-Excitation and Deep SupervisionforBrainTumorSegmentation,"IEEE,vol.8, pp.34029-34037,Feb2020.
[11] Q. S. W. WU DENG, "Deep LearningBased HCNN and CRF-RRNNModelforBrainTumorSegmentation,"IEEE, vol.8,pp.26665-26675,Jan2020
[12] S. R. TAMJID IMTIAZ, "Automated Brain Tumor Segmentation Based on Multi-Planar Superpixel Level FeaturesExtractedFrom3D MR Images,"IEEE,vol.8, pp.25335-25349,Dec2019.
[13] L.R.R.A.CHANGHEEHAN,"CombiningNoise-to-Image and Imageto-Image GANs: Brain MR Image Augmentation for Tumor Detection," IEEE, vol. 7, pp. 156966-156977,Sep2019.
[14] M.M.H.ABDUGUMAEI,"AHybridFeatureExtraction MethodWithRegularizedExtremeLearningMachinefor Brain Tumor Classification," IEEE, vol. 7, pp. 3626636273,Mar2019.
[15] Q.G.KaiHu,"BrainTumorSegmentationUsingMultiCascaded Convolutional Neural Networks and Conditional Random Field," IEEE, vol. 7, pp. 9261592629,Jul2019.
[16] W. A.-A. Nancy M. Salem, "MultiClassification of Brain TumorImagesUsingDeepNeuralNetwork,"IEEE,vol.7, pp.69215-69225,May2019.
[17] X.Y.AIMIN YANG,"ResearchonFeatureExtractionof TumorImageBasedonConvolutionalNeuralNetwork," IEEE,vol.7,pp.24204-24213,Feb2019.
[18] G.M.P.M.Shaker,"MachineLearningApproach-Based Gamma Distribution for Brain Tumor Detection and DataSampleImbalanceAnalysis,"IEEE,Nov2019.
[19] Z. G. Q. W. Qingneng Li, "Glioma Segmentation with a UnifiedAlgorithminMultimodalMRIImages,"IEEE,vol. 6,pp.9543-9553,Feb2018.
[20] W. L. ,. A. Z. Guotai Wang, "Interactive Medical Image SegmentationUsingDeepLearningWithImage-Specific FineTuning,"IEEE,vol.7,pp.1562-1573,Jan2018.
Volume: 09 Issue: 09 | Sep 2022 www.irjet.net p-ISSN: 2395-0072 © 2022, IRJET | Impact Factor value: 7.529 | ISO 9001:2008 Certified Journal | Page597