International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056

Volume: 09 Issue: 12 | Dec 2022 www.irjet.net p-ISSN: 2395-0072

International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056

Volume: 09 Issue: 12 | Dec 2022 www.irjet.net p-ISSN: 2395-0072
1Associate Professor, Dept. of Computer Science and Engineering, VNR Vignana Jyothi Institute of Engineering and Technology, Hyderabad, India 2, 3, 4, 5 Student, Dept. of Computer Science and Engineering, VNR Vignana Jyothi Institute of Engineering and Technology, Hyderabad, India ***
Abstract - Routine pathology workflows often involve a difference of opinion among pathologists and uncertainty of diagnosis. Deep learningshouldbe usedinthesesituations for enhancing the decision consistency and efficiency. Colon, StomachandKidneycancerareamongthehighestdeathrates nowadays. In this study, we propose a method to train a deep learning model to classify all gastric, colon and renal cancer using a single model. The Whole Slide Images are fed to the Efficient-Net model which is pre-trained on the ImageNet dataset. Themodelis trained usingatransferlearningmethod with partial transfusion. There have been various methods suggested to accurately classify these pathology images individually using partial transfusion. This work will demonstrate generalization ability of partial transfusion in classifying pathology images.
There have been many attempts at classifying the WSI imagesforindividualtissue[8,9,10].Weproposetocreatea generalizedmodelthatcanworkformanytissues,andfor this study we considered gastric, colon and renal tissues. Baqui,M.Netal.[5]carriedoutastudyatoneofthemost reputedhistopathologycentresinBangladesh.Theyfound that site of origin of tissue was absent in 71.83% of the containersamples.Thetissuenamewasalsoabsentin7.7% of the requisition papers. This information is critical for diagnosis and our model can be put into use in such situations.
Words: deep learning, pathology, image classification, transfer learning
Globalcancerstatisticsfromtheyear2022[1]digestive systemsitesstandasthesecondmostfrequentwith343,040 estimated cases right after genital system with 395,600 estimatedcases.Colonaloneaccountsfor106,180estimated newcasesinthedigestivesystem.Coloncancerisalsooneof the main causes of death in the modern world with a projection of 52,580 deaths in 2022. Stomach cancer accounts for 26,380 estimated cases and 11,090 deaths. Kidney cancer in the urinary system accounts for 79,000 estimatednewcasesand13,920deaths.
Whole slide images (WSIs), which are acquired by scanning glass slides with specialized equipment, are comparable to microscopy for primary diagnosis and providebenefitsofremoteconsultation.[2]Therehavebeen many applications proposed in computational pathology since the debut of deep learning. These techniques had successful results in applications like segmentation of tissues, classification, cell detection and many more [3,4]. Thesepromisingresultsshowthepowerofdeeplearningin pathology,andhowitcanavoidhumanerrorandeasethe clinicalworkflow.
One of the major challenges when using different datasetsisthevaryingimagesizes.So,wewouldfirstneedto performappropriatedatapre-processingtoscaletheimages tothesamesize.AndsincetheWSIimagesareverylarge,we willdividetheimagesintomanytilesusingfixedstridesand train the model using the individual tiles. We will use the partialtransferlearningmethod[6]tofinetuneapretrained model.Ourworkwilldemonstratewhetherpre-traineddeep learning models can be used with partial transfusion for generalizingpathologyclassificationfromanysiteoforigin.
Iizuka et al. [7] have classified gastric and colonic epithelialtumorsusingaweeklysupervisedmultipleinstance learning technique. After extracting many tiles from each WSIs,theytrainedCNNfromscratchusinginception-v3as the base architecture, reducing the number of weight parameters.Theymadeuseofslidingwindowtechniquewith stride256oninputtilesof512x512pixels,sinceallthetiles mustbeclassifiedtoobtaintheslidelabel. Themodelswere evaluated using max pooling and RNN (with two LSTM layers)aggregationmethods.Althoughtherewerealmostno statisticaldifferenceswiththemethodsusedforaggregation, if there was a difference RNN performed better. Adenocarcinoma classification got an AUC of 0.924 for stomachand0.982forcolon.Thelimitationofthisstudyis thatweneedahugedatasetforfollowingthismethodandit takesalongtimefortrainingamodelfromscratch.Anditis not worth the time when fine-tuned models also achieve similaraccuracyscores[6].
International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056

Volume: 09 Issue: 12 | Dec 2022 www.irjet.net p-ISSN: 2395-0072
Tsunekietal.[8]usedapartialtransferlearningmethod tofinetunethepre-trainedmodel.Theyhaveattemptedto classify the rare poorly differentiated colorectal adenocarcinomausingtheirmodel.Theirapproachmerely entailsmodifyingtheonlycertainparametersofthebatch normalization layers of the pre-trained model and not all. TheyusedtheEfficientNetB1modelwhichwastrainedon theImageNetdataset.The model wasabletoachieveROC AUCof0.94.
Byeon et al., [10] have used digital photographs of pathologyslidesratherthanusingWSIstoclassifycolorectal lesionsinto sixclasses.Theyachieved a mean accuracyof 97.3%usingDenseNet-161and95.9%usingEfficientNet-B7. Gradient-weightedclassactivationmappingwasutilizedin buildingasaliencymapthatshowstheareathatservedas the basis for the decision. An expert human pathologist assessedtheGrad-CAMpicturesofthetestdatasetandthe deeplearningmodel'sregionofinterestasacolormap.The limitationofthisstudywasthefactthatdigitalphotographs were used instead of the WSI images, since digital photographs are not an accepted standard for primary diagnosis.
BingluHuangetal.,[11]developeddeeplearningmodels thatcanpredictthediagnosisandcanalsofindtheoverall survivalofGastricCancerpatientsusingpathologicalimages. They developed two models using AI which are called GastroMILandMIL-GC.GastroMILmodelisfordiagnosing GastricCancerandMIL-GCforpredictingoutcomeofGastric Cancer.Theyachievedanaccuracyof92.0%thatisgreater than junior pathologist and also that is equal to senior pathologist.Fordevelopingthesemodels,theyincludedclear pathologicalimages,imagesofpatientswithGastricCancer andimagesofpatientsabove18years.Intheworkingmodel each image is divided into tiles with some size, then probabilityofthesetilesthatarebeingdiseasedisproduced asoutput.AfterthatKmostsuspicioustilesareselectedand finalpredictionoftheimageisgeneratedusingRNN.They also developed a working website for these models. The mainlimitationofthismodelissurvivalrateofGCpatientsis differentwithrespecttodifferentdatasetsduetoprogressof treatment.
Xiaodong Wang etal.,[12]developeda model thatcan predictcancerofthestomachfromaremovedlymphnode which contains three phases like Segmentation, Classification, T/MLN Ratio. To develop this model, they selected120WholeSlideImagesthatshowmetastasisfrom thetumorand60whole-slideimagesdevoidofmetastases. ToisolatetheareaofthelymphnodestheWSIslideisgiven asthesegmentationnetwork'sinputintheworkingmodel. The classification network then classifies tissues inside a lymphnodeareatodeterminethetumorregion.TheResNet50modelisusedforclassificationwhichhasmoreaccuracy andinferencespeedthanInceptionV4andalsoResNet-101.
This method's main flaw is that it used a dual-center retrospective research of GC from one country for its prognosticanalysisofT/MLN.
HishamAbdeltawabetal.,[13]suggesteda newmodel forautomatedclassificationtotakecareofdiagnosticchores using a deep learning pipeline. This approach can both identify and categorize kidney tumor- and non-tumorbearingregions.TheframeworkconsistsofthreeCNNsand WSIimagesofthekidneythataredividedintogroupsof3 differentsizeswhichareconsideredasinput.Thismodelcan provide patch wise and pixel wise classification. The suggestedmodelhasa92.0%accuracyrate.Thepyramidal deeplearningmodelthathasbeenpresentedmakesuseofa hierarchyofthreeCNNstohandlevariouspicturesizes.In thismodeltheimagesizesare250*250,350*350,450*450. The main limitation of this model is mislabeled images, foldedtissuesandtorntissues.
Yasmine Abu Haeyeh et al., [14] For RCC subtyping, a multiscaleweaklysuperviseddeep-learningtechniquewas developed.TheRGB-Histogramstandardstainnormalization isfirstappliedtotheentire slidepicturesintheproposed systemtoreducetheimpactofcolorfluctuationsonsystem performance. To retain the tissue connectedness, they dividedtheinputdataintoseveraloverlappingpatchesusing the multiple instance learning technique. To get the final classificationjudgment,theytrainthreemultiscaleCNNsand then apply decision fusion to their anticipated outcomes. With a 97.9% accuracy rate and no need for fine-grained annotationsatthepatchlevel,thismodeleasestheloadof pathologists'annotations.
MichaelFenstermakeretal.,[15]createdadeeplearning model to determine whether histopathological specimens include Renal Cell Carcinoma (RCC), which had a 99.1% accuracy rate. To maintain the RGB color channel information,adigitized(H&E)slideisdividedintogroups andrepresentedasa3Dmatrixinthisapproach.Onepattern issoughtafterbyafeaturedetector.Inordertobeanalyzed byaneuralnetwork,thepooled2Dpictureisflattenedtoa 1D vector. The data is flattened and sent through several layersofneurons.thatareallcompletelyconnected,upuntil thelayerthatdeterminessubclassprobabilitiesinthefinal output layer. In this model, the algorithm successfully identifiedtherelatedultimatepathologyinmorethan97% ofinstances,evenwhenjustusingasingle100um2patchof renaltissue.
Amit Kumar Chanchal et al., [16] proposed a utilizing deeplearningarchitectureSeparableConvolutionPyramid Pooling Network (SCPP) for segmentation of histopathological images. They worked on three datasets that are breast cancer, kidney, multi organ disease histopathology.Encoder-decoderarchitectureservesasthe foundationforsegmentation,SCPPmorepertinentfeatures
International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056

Volume: 09 Issue: 12 | Dec 2022 www.irjet.net p-ISSN: 2395-0072
are being extracted at a higher level. When compared to earlier deep learning models that had less computational complexity,thesuggestedSCPP-Netarchitectureperformed thebestatF1andAJIscore.
Glovanna Maria Dimitri et al., [17] suggested a solid decision-making aid for the automated assessment of the Karpinskimeasure.Thiswillassistmedicalprofessionalsin determiningwhetherornotthekidneyistransplantableby assisting them in identifying the presence of sclerotic glomeruli.UsinghumankidneysamplesfromscannedWhole Side Images, sclerotic and non-sclerotic glomeruli were identified and distinguished. The DeepLab V2 model was usedtosegmentthepictures,and512×512patchestaken from the original WSIs were encoded using a pretrained ResNet101encoder.
Fengyi Li et al., [18] suggested a unique technique for fine-grainedglomerulicategorization.Glomeruliwasdivided asNeg-tubuleandartery,SS-anareaoftheglomerulartuft thathassclerosis,GS - sclerosis affectingtheentiretuft in full, C - Bowman's space has more than two layers of collectingcells,usuallywithdepositsofcollagenandfibrin; NOA-noneoftheabove.DenseNet-121choosestheGSand Negsamples,whicharethenforwardedtotheglomerular refinement module. A component of the adversarial correlation guided network is the feature extractor, along withaseparatorandanadversarialcorrelationloss(CGN).
S. Shubham et al., [19] locating human kidney tissues usingdeeplearningtoidentifyglomeruli.Inordertoscale several dimensions utilizing a primary technique, UNet is using EfficientNetB4 as the segmentation model, and foundation.ThedatasetusedwasHumanBioMolecularAtlas Program (HuBMAP). When the area of interest (ROI) in medicalimagingisambiguous,itisfeasibletoovercomethe constraintsofUNetdeepneuralnetworkbyfine-tuningthe model.
Fuzhe Ma et al. [20] In order to identify and segment chronic renal failure, an HMANN model (Heterogeneous ModifiedArtificialNeuralNetwork)thatissupportedbya Backpropagation(BP)approachandclassifiedasaSupport Vector Machine (SVM) and Multi-Layer Perceptron (MLP) wascreated.Thepre-processingstepsinvolvedrestoringof the image, sharpening and smoothening, contrast enhancementmethodwhichhelpedindenoising.Theabove processwasabletoachievehighaccuracyandperformance byincludinglevelsetsegmentationduringdenoisingofthe images.
J.Yogapriyaetal.[21]havedoneacomparativeanalysis by considering some of the pre-trained models like GoogleNet,ResNet-18,VGG16,aCNNmodel(Convolutional NeuralNetwork).ThedatasetusedareGIT(GastroIntestinal Tract) images from Norway’s VV health trust taken with
endoscopic equipment. The small data sizes issue was resolved by using transfer learning to fine-tune the developedmodel.Theanalysismadeitclearthat,fromthe pre-trainedmodelsconsidered,theVGG16modelwasableto achieve betterperformance with96.33% accuracy,96.5% precision, 96.5% F1-measure, and 96.37% recall. The requirementofmanuallyupdatingtheinformationwhichis then used as the dataset, is the weakest point of the algorithm. By using larger datasets, the above-mentioned issuecanbesolved.
DeivaNayagametal.[22]haverecommendedutilizing digitizedH&Estainedhistologyslidestododeeplearningto identifycolorectalcancer.Heclaimsthat,whencomparedto all other methods and techniques, the CNN model's classification of images of colon cancer tissues has the highest accuracy and the shortest computation time. Accuracy of 99.7% was achieved using CNN as the main model. The model provides a 99.9% median accuracy for normalslidesand94.8%medianaccuracyforcancerslides. Themodel’spredictionswerecomparativelybetterthanthat of a pathologist. Use of supervised techniques for image classificationoutperformedtheunsupervisedtechniques.
EiichiroUchinoetal.[23]focusedprimarilyontheseven pathological characteristics necessary for a diagnosis, including segmental sclerosis, crescent, endocapillary proliferation,globalsclerosis,andstructuralmodificationsto thebasementmembraneandcrescent.Usingtheinformation mentioned,anAIbasedmodelwasdevelopedtoclassifythe abovefindings.Thedatasetusedforthismodeldevelopment consistedoftheWSIsfrom283renal biopsypatientswho had their procedures approved by the Kyoto University Hospitalforresearchuse.Themodelperformedwell,with anAUCofmorethan0.98,whichwasvirtuallyidenticalto nephrologists'results.Aweakpointofthemodelcouldbe thatitbecomestoughtopredictaccuratelywhentheinput deviates from the data largely. Another limitation of this modelisthatthedatasetconsideredwereinvolvingonlyPAS andPAMstaining.
Inthisstudy,weconsideredthreedifferentdatasets,firstis Dartmouth Kidney Cancer Histology Dataset, second is HistopathologicalimagesofColonandLungCancerandthird is Gastric Slice Dataset. The detailed description of these datasetsisgivenbelow.
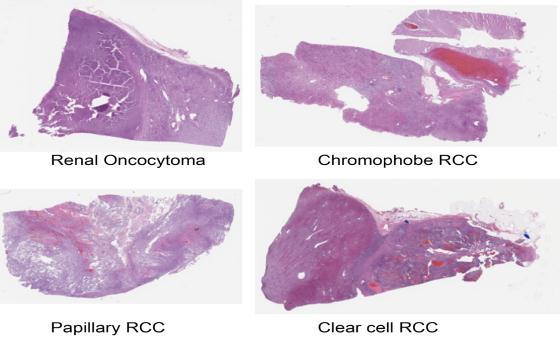
ThePathologyandLaboratoryMedicineDepartmentat Dartmouth-Hitchcock provided this dataset of 563 wholeslidepicturesofrenalcellcarcinoma(RCC)Formalin-fixed paraffinembedded(FFPE)andhematoxylinandeosin(H&E) stainedimages(DHMC).
International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056

Volume: 09 Issue: 12 | Dec 2022 www.irjet.net p-ISSN: 2395-0072
Sinceweareusingrelativelysmalldatasetstohavearobust performance,wewillfirstapplydataaugmentationonthe datasetsandcombinethem.SincetheWSIimagesarevery largewewillperformslidetilingbyextractingsquaretiles withafixedstridefromtissueregionsandtraversethemina slidingwindowfashionwhiletrainingthemodel.
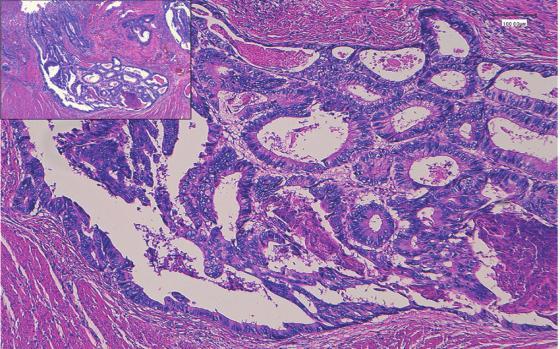
Histopathological Pictures of Lung and Colon CancerThis collection includes 25,000 histopathological images dividedinto5groups.JPEGfileswitharesolutionof768by 768 pixels make up each image. The original sample of verified sources that complied with HIPAA was used to createthe500photosofcolontissueand750imagesoflung tissue(including250eachofbenign,adenocarcinoma,and squamouscellcarcinomas)(250benigncolontissueand250 colonadenocarcinomas).TheAugmentor programme was thenusedtoenhancethesephotosto25,000.
We will use Transfer Learning on EfficientNetB7 architecture[24]andfinetuneittoourdataset.EfficientNet offersCompoundScalingwhichcombinestheDepth,Width and Resolution Scaling in an efficient manner providing a modelwithsmallersizeandhigheraccuracy.Thereasonfor usingapretrainedmodelinsteadoftrainingamodelfrom scratch is the faster training time with a similar model performance.
We will take advantage of Partial Transfusion [6] and fine-tune only the trainable weights of the batch normalizationlayersasitleadstosimilarperformanceasto fine-tuningalloftheweightsalongwithfastertraining.This meansthatwecanonlytraintheparametersscaleandoffset, insteadofalltheparametersandreducetheoveralltraining timeonceagain.ThemodelwillbetrainedwiththeAdam optimisationalgorithm[25]andcrossentropylossfunction.
In order to perform transfer learning with the given modelfewchangesneedtobemadetothepretrainedmodel. First,weneedto remove thefinal classificationlayerasit outputsthelabelsfortheImageNetdataset.Thenweapplya Global-Average-Pooling(GAP)layerfollowedbyadropout layer. A softmax activated dense layer at the end will be addedtoclassifyintoourtargetlabels.
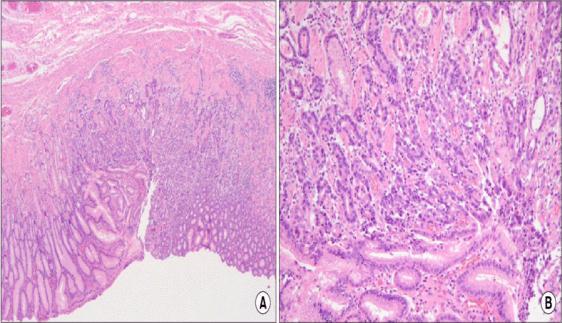
GastricSliceDataset-Thegastricslicedatasetcontains 140normalpicturesand560imagesofmalignancy.
In this study, we have done a lot of research work about pathology classification, we have explored numerous researchpapersrelatedtopathologyanditsstatistics.We came to know that among various diseases the cancers related to the digestive system are some of the most frequent, especially gastriccancer, colon cancer and renal cancer. Pathologists find it very difficult to detect these diseasesmanuallywithexistingsystems.Therefore,alotof work needs to be done to help pathologists to reduce the time-consuming workflow of identifying diseases and diagnosingthedisease.Furthermore,wehavelearnedabout various existing systems and proposed systems in the research papers which helps us in building our new proposedmodelthathelpspathologiststodetectthedisease veryquicklyandeasily.
International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056

Volume: 09 Issue: 12 | Dec 2022 www.irjet.net p-ISSN: 2395-0072
Specialthankstoourteamguide,Dr.P.V.SivaKumar,forall the technical support and guidance which led to the completionoftheliteraturesurveyphaseoftheprojectwith agoodresultintheend.
[1] Siegel RL, Miller KD, Fuchs HE, Jemal A. “Cancer statistics,2022”.CACancerJClin.2022;72(1):7–33.
[2] ShinoharaS,BychkovA,MunkhdelgerJ,KurodaK,Yoon HS, Fujimura S, Tabata K, et al. “Substantial improvementofhistopathologicaldiagnosisbywholeslideimage-basedremoteconsultation”.VirchowsArch. 2022Aug;481(2):295-305. doi:10.1007/s00428-02203327-2. Epub 2022 Jun 7. PMID: 35672584; PMCID: PMC9172976.
[3] Chetan L. Srinidhi, Ozan Ciga, Anne L. Martel, “Deep neural network models for computational histopathology: A survey”, Medical Image Analysis (2020), doi: https://doi.org/10.1016/j.media.2020.101813
[4] VanderLaak,J.,Litjens,G.&Ciompi,F.“Deeplearningin histopathology:thepathtotheclinic”.NatMed27,775–784 (2021). https://doi.org/10.1038/s41591-02101343-4
[5] Baqui, M. N., Rozhana, S., Rajib, R. C., Huda, M. M., Mohammad Mesbahuzzaman, .-., & Paul, T. K. (2019). “FrequencyofMislabeledSpecimeninaHistopathology Laboratory”.JournalofSurgicalSciences,17(2),80–83. https://doi.org/10.3329/jss.v17i2.43726
[6] Kanavati, F.; Tsuneki, M. Partial transfusion: “On the expressiveinfluenceoftrainablebatchnormparameters fortransferlearning”.arXiv2021,arXiv:2102.05543.
[7] Iizuka, O., Kanavati, F., Kato, K. et al. “Deep Learning Models for Histopathological Classification of Gastric andColonicEpithelialTumors”.SciRep10,1504(2020). https://doi.org/10.1038/s41598-020-58467-9
[8] Tsuneki, M.; Kanavati, F. “Deep Learning Models for Poorly Differentiated Colorectal Adenocarcinoma Classification in Whole Slide Images Using Transfer Learning”. Diagnostics 2021, 11, 2074. https://doi.org/10.3390/diagnostics11112074
[9] Ahmad,N.,Asghar,S.&Gillani,S.A.“Transferlearningassisted multi-resolution breast cancer histopathologicalimagesclassification”.VisComput38, 2751–2770 (2022). https://doi.org/10.1007/s00371021-02153-y
[10] Byeon, Sj., Park, J., Cho, Y.A. et al. “Automated histologicalclassificationfordigitalpathologyimagesof colonoscopy specimen via deep learning”. Sci Rep 12, 12804 (2022). https://doi.org/10.1038/s41598-02216885-x
[11] BingluHuang,ShanTian,NaZhan,JingjingMa,ZhiweiHuang, ChukangZhang,HaoZhang,FanhuaMing,FeiLiao,MengyaoJ i,JixiangZhang,YinghuiLiu,PengzhanHe,BeiyingDeng,Jiam ingHu,WeiguoDong:“Accuratediagnosisandprognosis prediction of gastric cancer using deep learning on digitalpathologicalimages:Aretrospectivemulticentre study”
[12] XiaodongWang,YingChen,YunshuGao,HuiqingZhang, Zehui Guan, Zhou Dong, Yuxuan Zheng, Jiarui Jiang, HaoqingYang, LimingWang,XianmingHuang,LirongAi,WenlongYu, HongweiLi,ChangshengDong,ZhouZhou,XiyangLiu& GuanzhenYu:“Predictinggastriccanceroutcomefrom resectedlymphnodehistopathologyimagesusingdeep learning”https://doi.org/10.1038/s41467-021-216747
[13] HishamAbdeltawab,FahmiKhalifa,MohammedGhazal, Liang Cheng, Dibson Gondim & Ayman El-Baz: “A pyramidaldeeplearningpipelineforkidneywhole-slide histology images classification” https://doi.org/10.1038/s41598-021-99735-6
[14] Yasmine Abu Haeyeh, Mohammed Ghazal, Ayman ElBaz,ImanM.Talaat:“DevelopmentandEvaluationofa Novel Deep-Learning-Based Framework for the Classification of Renal Histopathology Images” https://doi.org/10.3390/bioengineering9090423
[15] MichaelFenstermaker,ScottA.Tomlins,KarandeepSingh, JennaWiens, Todd M.Morgan: “Development and ValidationofaDeep-learningModeltoAssistWithRenal Cell Carcinoma Histopathologic Interpretation” https://doi.org/10.1016/j.urology.2020.05.094
[16] Amit Kumar Chanchal, Aman Kumar, Shyam Lal, Jyoti Kini:“Efficientandrobustdeeplearningarchitecturefor segmentation of kidney and breast histopathology images” https://doi.org/10.1016/j.compeleceng.2021.107177
[17] Giovanna Maria Dimitri , Paolo Andreini, Simone Bonechi,MonicaBianchini,AlessandroMecocci,Franco Scarselli,AlbertoZacchiGuidoGarosi,ThomasMarcuzzo andSergioAntonioTripodi,“DeepLearningApproaches for the Segmentation of Glomeruli in Kidney Histopathological Images” https://doi.org/10.3390/math10111934
International Research Journal of Engineering and Technology (IRJET) e-ISSN: 2395-0056

Volume: 09 Issue: 12 | Dec 2022 www.irjet.net p-ISSN: 2395-0072
[18] F. Li et al., "Correlation-Guided Network for FineGrainedClassificationofGlomerularlesionsinKidney Histopathology Images," 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), 2020, pp. 57815784,doi:10.1109/EMBC44109.2020.9176234.
[19] Shubham,S.,Jain,N.,Gupta,V. et al. “Identifyglomeruli in human kidney tissue images using a deep learning approach”. Soft Comput (2021). https://doi.org/10.1007/s00500-021-06143-z
[20] FuzheMa,TaoSun,LingyunLiu,HongyuJing,“Detection and diagnosis of chronic kidney disease using deep learning-basedheterogeneousmodifiedartificialneural network”, Future Generation Computer Systems, Volume 111, 2020, Pages 17-26, ISSN 0167-739X. https://doi.org/10.1016/j.future.2020.04.036.
[21] J. Yogapriya, Venkatesan Chandran, M. G. Sumithra, P. Anitha, P. Jenopaul, C. Suresh Gnana Dhas, "Gastrointestinal Tract Disease Classification from Wireless Endoscopy Images Using Pre-trained Deep Learning Model", Computational and Mathematical MethodsinMedicine,vol.2021,ArticleID5940433,12 pages,2021.https://doi.org/10.1155/2021/5940433
[22] DeivaNayagamR,AarthiK,MirraS,2022,“ColonCancer Classificationon Histopathological Imagesusing Deep Learning Techniques”, INTERNATIONAL JOURNAL OF ENGINEERING RESEARCH & TECHNOLOGY (IJERT) ETEDM–2022(Volume10–Issue08).
[23] Eiichiro Uchino, Kanata Suzuki, Noriaki Sato, Ryosuke Kojima, Yoshinori Tamada, Shusuke Hiragi, Hideki Yokoi,NobuhiroYugami,SachikoMinamiguchi,Hironori Haga,MotokoYanagita,YasushiOkuno,“Classificationof glomerular pathological findings using deep learning and nephrologist–AI collective intelligence approach”, International Journal of Medical Informatics, Volume 141, 2020, 104231, ISSN 1386-5056, https://doi.org/10.1016/j.ijmedinf.2020.104231
[24] Tan,M.;Le,Q.“Efficientnet:Rethinkingmodelscalingfor convolutional neuralnetworks”.InProceedingsofthe International Conference on Machine Learning, Long Beach,CA,USA,9–15June2019;pp.6105–6114.
[25] Kingma, D.P.; Ba, J. Adam: “A method for stochastic optimization”.arXiv2014,arXiv:1412.6980.
[26] Song, Z., Zou, S., Zhou, W. et al. Clinically applicable histopathological diagnosis system for gastric cancer detection using deep learning. Nat Commun 11, 4294 (2020).https://doi.org/10.1038/s41467-020-18147-8