e-ISSN:2395-0056
Volume: 09 Issue: 10 | Oct 2022 www.irjet.net p-ISSN:2395-0072


e-ISSN:2395-0056
Volume: 09 Issue: 10 | Oct 2022 www.irjet.net p-ISSN:2395-0072

Yash Thakre1 , Yugantkumar Gajera2 , Shrushti Joshi3, Jose George4
1National Institute Of Technology, Raipur, India
2Bhagwan Arihant Institute Of Technology, Surat, India
3Jai Hind College, Mumbai, India
4Niagara College of Applied Arts and Technology, Ontario, Canada ***
Abstract - Medical image processing using machine learning is an emerging field of study which involves making use of medical image data and drawing valuable inferences out of them. Segmentation of any body of interest from a medical image can be done automatically using machine learning algorithms. Deep learning has been proven effective in the segmentation of any entity of interest from its surroundings such as brain tumors, lesions, cysts, etc which helps doctors diagnose several diseases. In several medical image segmentation tasks, the U-Net model achieved impressive performance. In this study, first, we discuss how a Dilated Inception U-Net model is employed to effectively generate feature sets over a broad region on the input in order to segment the compactly packed and clustered nuclei in the Molecular Nuclei Segmentation dataset that contains H&E histopathologypictures, includinga comprehensivereview ofrelatedworkontheMoNuSegdataset.
Key Words: Dilated Inception U-Net, Nuclei Segmentation, Dilated Convolutions, Convolutional Neural Networks, Deep Learning
The methods of prognosis and prediction of cancer in patients have been improving and being researched consistently over the past years. Predicting cancer susceptibility, predicting cancer resurgence, and forecasting cancer survivability are the three areas of interestwhilepredictingcancer[1]. Itisvitaltosegment critical organs, tissues, or lesions from medical images andto extract features from segmentedobjectstoassist physicians in making the correct diagnosis. Cancer detection in patients is made easier using machine learning algorithms such as Artificial Neural Networks, Decision Trees, Support Vector Machines, and other classificationalgorithmsthatlearnfeaturesandpatterns from the past patient data provided to them and make predictions on the unseen data using the learned featuresandpatternsfromthepasttruedata.
Segmentationofanobjectfromamedicalimagehas been an intricate task in medical image analysis which can be realized using machine learning models [2] instead of manual annotation by hand. This is usually accomplished by feeding a 2-dimensional or 3dimensional image to a machine learning algorithm and acquiring a pixel-wise classification of the image as a prediction. Deep learning, an emerging field of machine learning, turns the original feature representation space into another space by layering feature transformations, making tasks like recognition, classification, and segmentation easier which is achieved by using convolutional neural networks that look for valuable patterns in the image. This method of learning samples using big amounts of medical data can better characterize the rich information inherent in the data thantypicalartificialmethodsforconstructingfeatures.
Nuclei segmentation in histology images helps doctorsdiagnosecancerinpatients.Thenuclei'scontour andsizearethemostsignificantfeaturesthatneedtobe predicted in a medical image for an appropriate diagnosis. Therefore, modifications to the original DIUNet model are proposed to segment the nuclei in the MoNuSeg data [3]. ADilated InceptionU-Netmodel was usedwhichusesdilatedconvolutionsthatarecapableof efficiently generating feature sets of a large area on the input. The model proposed was much more computationallyefficientandfocusedoncapturingmore details in the data as compared to other reviewed models in the same sphere. This was done by introducing dilated inception blocks instead of the traditionally used convolutional layers to overcome the shortcomings of classic U-Net on the MoNuSeg dataset. Thesedilationsenabledthemodeltolearnfeaturesfrom a larger spatial domain without being very computationally expensive. The dataset used for testing theproposedmodel wasintroducedbyNeerajKumaret al2017asapartoftheMulti-OrganNucleiSegmentation (MoNuSeg) Challenge [4]. The data includes images of manually annotated, magnified nuclei that are
International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056

hematoxylin and eosin (H&E) stained, which are the bodiesofinterestthatneedtobesegmented.
Advances in Deep neural networks in medical imaging have been used extensively for localization and classification of nuclei in histopathology data from breast and colon cancer, however, such traditional methods did not work for segmentation Thus this introduced the use of Deep CNN architectures for semantic segmentation due to its automatic feature extraction and end-t-end training methods Then came Fully Convolutional Networks that could be applied regardless of input image sizes and produce more accurate segmentation results by utilizing features from different scales [5] Traditional segmentation strategies likethe SegNet [6] and the classicU-Netmodel [7] keep encodingtheimagetoabottlenecklayertoextractmore influential features from the image and using skip connectionstoretainthespatialfeaturestosegmentthe object of interest from the surrounding, have shown good results but have struggled to delineate nucleus borders adequately. Specifically designed models have been introduced recently to segment overlapped and clusterednucleifromthisdataset.
Z Zeng et al 2018 introduced a RIC U-Net model which comprises deep residual inception modules and residual modules along with convolutional layers as the skip connections of the U-Net model and segmented the clustered/overlapped nuclei from the same histology images in the MoNuSeg dataset [8]. They used an attention mechanism in the decoder blocks to select the mostinfluentialfeatures.However,thenetwork wastoo deepandthenumberofimagesprovedtobeinsufficient which led the model to overfit. Zhou et al 2020 used a multi-Head[9] FullyConvolutionalNetworkmodelwith a ResNetwithfifty convolutional layersasthebackbone for the top-down encoder and a binary cross-entropy loss function to segment the nuclei in this dataset. They also normalized the color of the tissue images and dilated the binary masks once before using them as the training data. The patch sizes used by them were 256 x 256.
Tahir Mahmood et al 2021 proposed a nuclear segmentation method based on a residual skip connection It did not require post processing unique traditional nuclei segmentation strategies They emphasized on utilizing residual connectivity to maintain the information transfer from the encoder to thedecoderTheyusedthestainnormalizationtechnique asproposedbyMacenko [10].TheyproposedanR-SNN, an end-to-end encoder–decoder segmentation network
in which the input image is first downsampled by passing it through multiple deep-learning convolution andpoolinglayersintheencoderandthenupsampledto the original size by the decoder part. They used crossentropy loss because of its logarithmic function and probabilisticapproach.Theyutilizedstainnormalization toreducethenumberofconvolutionlayersandthusthe proposed method had fewer trainable parameters, the model converged rapidly and trained fast. Kiran I et al 2022segmentedtheseclusterednucleibyadoptingpreprocessing techniques [11] like color normalization, patch extraction, data augmentation, distance mapping, and binary thresholding by introducing a DenseRes UNet model which replaced the skip connection in U-Net with atrous blocks (or dilated convolutions) to ensure thereisnodimensionmismatchingbetweentheencoder blocks and decoder blocks. The authors performed distancemappingtofindthenuclei'scenterpointandto distinguishbetweentheinnerboundaryandcoreareaof the nuclei. Binary thresholding was applied to the distance maps before feeding them to the DenseRes UNet model. Yunzhi Wang cascaded two U-Nets together toconstructamodelandappliedcolornormalizationon thenucleiimagesusingthemeanandstandarddeviation from the ImageNet dataset [12]. The author used 512x512 patches for training, and patches of this size makethemodeltrainrelativelymoreslowly.KongYetal 2020 Two-Stage Stacked U-Nets with an attention mechanismthatusesU-Netasthebackbonearchitecture andinputimagesonfourdifferentzoomedscalesandan attention generation model which is used to weigh the outputs of these four differently scaled sets of inputs[13]. They predicted the masks in the two stages, but also fed the input along with the predicted masks from the first stage to the second stage, attaining the finalprediction.
Excessive data augmentation and pre-processing measures on the dataset have certainly helped deep learning models generalize well on unseen data. Color normalizationhasbeenacommonstrategytoovercome the influence of stain variations in the dataset on the models. These methods avoid the potential model overfittingtothetrainingdata.Themodelmakinguseof dilated convolutions has substantially less trainable parameters as compared to the models that extract featuresetsofthesamescale.
Volume: 09 Issue: 10 | Oct 2022 www.irjet.net p-ISSN:2395-0072 © 2022, IRJET | Impact Factor value: 7.529 | ISO 9001:2008 Certified Journal | Page 843
D Cahall et al 2017 introduced Dilated Inception UNet for brain tumor segmentation on the BRATS 2018 dataset and achieved impressive results [14, 15]. However,theMoNuSegdatasetwascomparativelymore complexandcontainedclusterednuclei.Tobetterfitthis
International Research Journal of Engineering and Technology (IRJET)

e-ISSN:2395-0056
Volume: 09 Issue: 10 | Oct 2022 www.irjet.net p-ISSN:2395-0072
data, a few changes were made to the Dilated Inception U-Net model to use for the segmentation of nuclei. The challenge was not only to segment the nuclei but to eliminatetheoverlappingnucleiinthepredictedmasks.
The DIU-Net model for nuclei segmentation was implementedonthetrainingdataalongwithaugmented data to segment the nuclei on the unseen testing data. The dilated Inception U-Net model is able to extract a diverse set of features that include more spatial information from the tissue images. The preparation of training the data commenced with the extraction of patches from the high-resolution histology images followed by the application of a few data augmentation techniques to compensate for the insufficient training dataavailable.
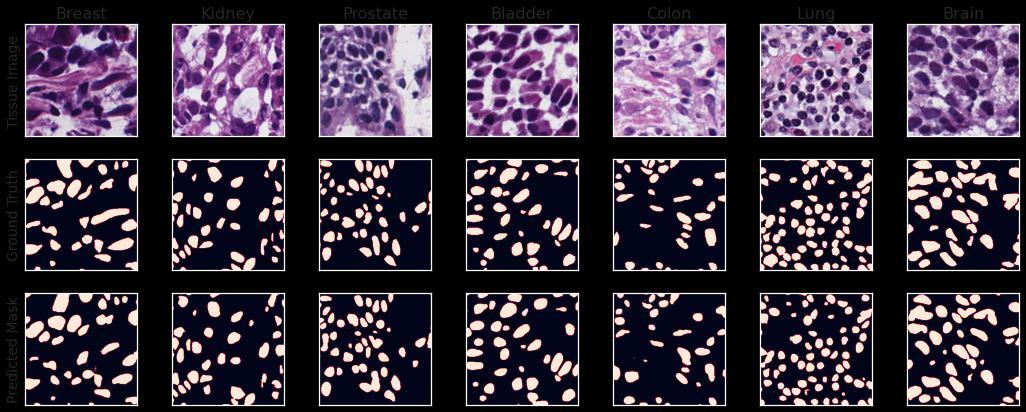
The dataset consists of 30 Hematoxylin and Eosin (H&E) stained tissue images with more than 21000 nuclear boundary annotations validated by a medical doctor.Thedatasetwasintroducedby(NeerajKumaret. al. 2017) who generated this dataset from The Cancer Genomic Atlas (TCGA) archive [16]. These 30 images contained H&E-stained images of 7 organs (Bladder, Brain, Breast, Colon, Kidney, Lung, and Prostate) which were used for training the model. The corresponding masks to these images were in the form of XML files, so theywerefirstconvertedtobinarymasksbeforefurther operations. Additionally, 14 images were provided to evaluate the model's performance and were utilized to validateandevaluatethemodel.
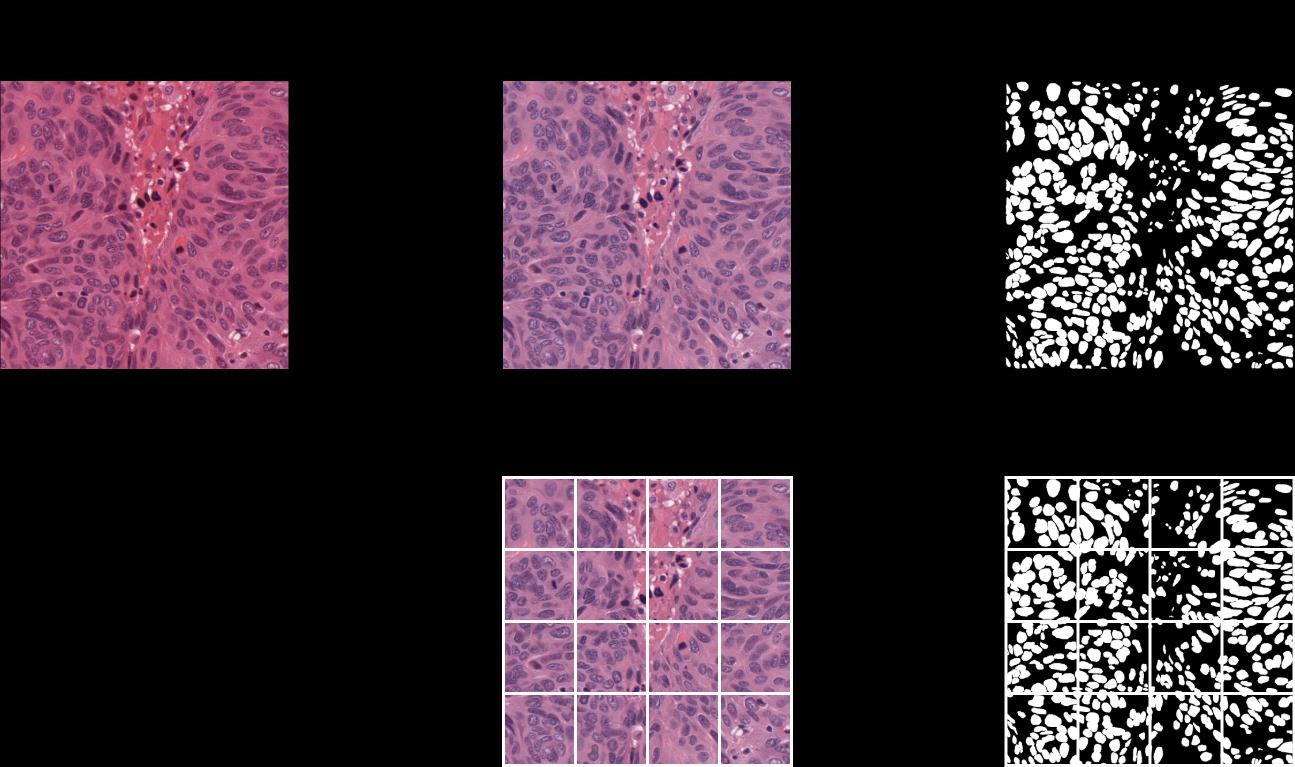
In order to tackle this difficult dataset, a few preprocessing techniques for the data were adopted, including patch extraction from raw images to reduce the load while training the model, and augmentation to deal with the shortage of data for training a Convolutional Neural Network model. The H & E stain variations across the organs pose threats to the quality of model training. To avoid this, a color normalization technique was adopted which involves the Singular Value Decomposition geodesic method for the acquisitionofstainvectors oftheimages [10].Thecolor normalized images were then used for further preprocessing. To prepare the training data, patches of dimensions 256 x 256 were extracted from every training and testing image of dimensions 1000 x 1000 with some overlapping (Fig 1). Each image would provide 16 patches with a small overlapping. The corresponding masks were also patched the same way before feeding to the model. The training patches and testing patches were kept separate throughout the training. To avoid potential overfitting of the model to the training data, multiple augmentation techniques were applied including random rotation, vertical flip, horizontalflip,gaussianblur,gaussiannoise,colorjitter, and channel shuffle. The open-source computer vision library OpenCV and TensorFlow were used to apply theseaugmentationtechniquestotheimagepatches[17, 18]. Besides the 480 training images, there were approximately 2500 augmented images that were used along with the unaugmented data to train the model, keepingthepatchesfromthe14imagesfromthetesting dataasideasvalidationdata.
International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056

Volume: 09 Issue: 10 | Oct 2022 www.irjet.net p-ISSN:2395-0072
1.
The classic U-Net model was modified by applying dilated inception blocks instead of the traditional convolutionlayerblocks.Theprimarygoalistoperform classificationonallthepixelsofthepatchesofthetissue imageandproducea binarymaskthatshowswhethera pixel belongs to a nucleus or not. Every pixel in the binary mask has either two values 0 and 1 after binary thresholdingontheoutputfromthefinalsigmoidlayer.
Dilated filters in addition to standard filters were employed to capture features from various spatial domains and create a feature set from these combined sets of features. The dilated filters capture just as much information as the standard filters capturing a larger areahavingthesamenumberofparameters.Therewere significantly fewer trainable parameters in the model, which significantly reduced the load caused by the model's training. Inception modules have been effective in extracting significant features from images that help in image classification [19]. Due to their variable filter sizes in the inception layers, the network can learn
variousspatialpatternsatdifferentscales.Consequently, the numberof trainablefiltersisincreasedsubstantially if the same number of filters are used. Dilated filters solve this shortcoming by detecting similar spatial patterns on the same scale with less trainable parameters.
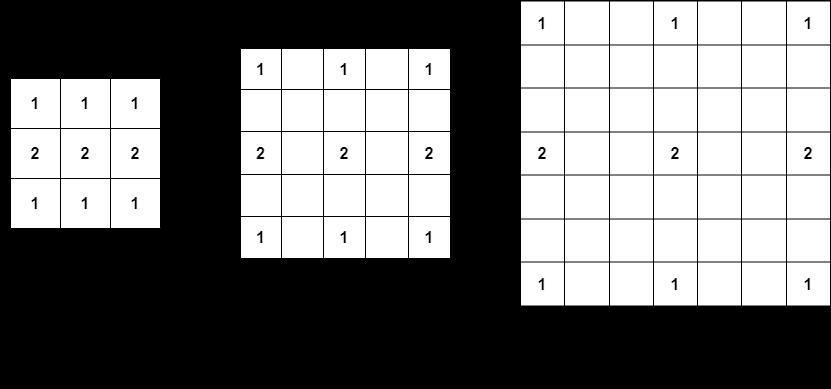
The purpose of dilated convolutions is to gather information over large areas of the image. Dilation operates as if the filter is expanded and zeros are introduced in between the gaps and those zeros are phantoms i.e., not trainable. In other words, these dilationsgrowtheregioncoveredbyafilter.Fig2shows how these dilations affect the filter’s region using an example. The underlying motivation behind using dilated filters is to capture information from a larger area without dealing with too many trainable parameters.
International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056

Volume: 09 Issue: 10 | Oct 2022 www.irjet.net p-ISSN:2395-0072
Fig 2. Aconvolutionfilterwithdifferentdilationrates.
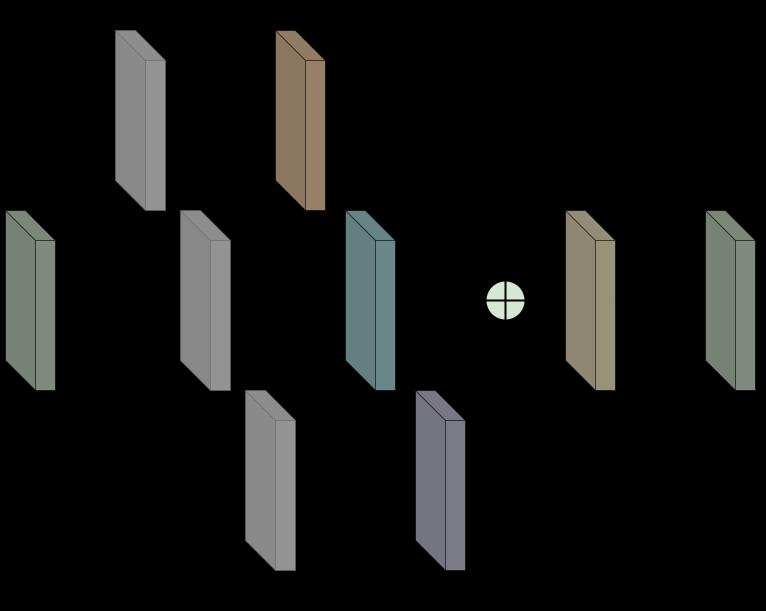
Input is followed by three parallel 1x1 filtered layers to ensure that the following three parallel differently dilated layers of 3x3 filters receive input with significantly fewer channels to reduce the load while training. The differently dilated filters use same zeropadding which means there are a sufficient number of zeros introduced to the edges of the input to make sure theoutputhasthesamesizeastheinputtoconcatenate the three outputs from the three differently dilated convolution layers. All the filters in these blocks are activatedbyaRectifiedLinearUnitfunction(ReLU). The 1x1filtersareusedinordertoreducethedimensionsof the input which would lower the number of trainable
parameters. The outputs from the three differently dilatedfiltersareconcatenatedtogetherandfollowedby a batch normalization layer which produces the output from each of these blocks (Fig 3). The batch normalization layer is used to normalize the output of the preceding layers, making learning more effective. It can also be used as regularization to prevent the model fromoverfitting.Thenaboveeachconvolutionallayerin the block represents the number of filters in that layer andthis number can beusedto calculatethe number of channels in the output of each block (i.e., 3n for a concatenation of three outputs of n channels each). TheseblocksarethebasicunitsfortheDilatedInception U-Netmodel.
International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056

Volume: 09 Issue: 10 | Oct 2022 www.irjet.net p-ISSN:2395-0072
Fig 3. DilatedInceptionBlock
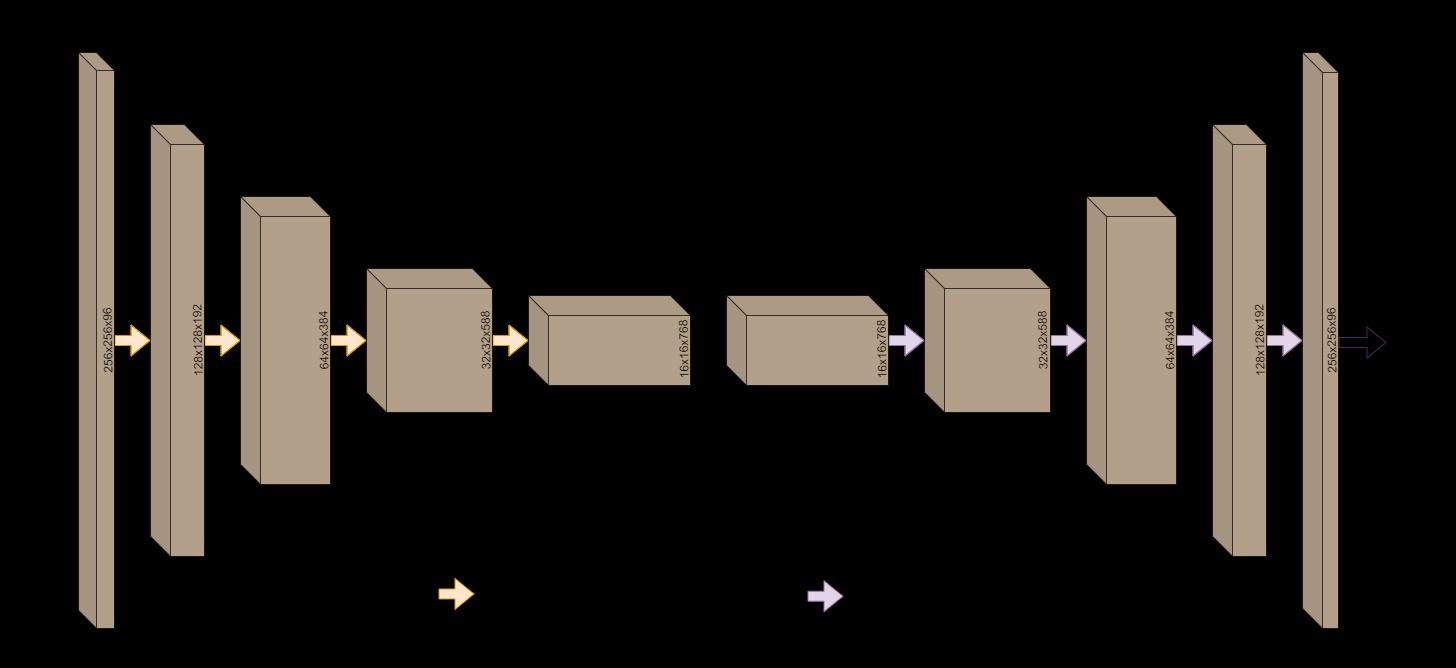
Thedilatedinceptionblocksareputtogethertoform the dilated inception U-Net model, which modifies the originalU-Netmodelforsegmentation.Fig4displaysthe model architecture constructed by the dilated inception blocksinsidewhichcanbeseeninFig3.Theshapeofthe outputs from each of the blocks has been written on them. These blocks are used to extract more spatial information on each encoding and decoding block as well as the two bottleneck blocks. The purpose of this
model is to extract useful features from the input image while keeping the spatial information about the image intact. A sigmoid activation layer is used finally to classifywhetherapixelbelongstoanucleusinthetissue image by mapping the output from the last block to valuesintherange0to1.Closervaluesto1indicatethe presenceofapixelbelongingtoanucleus.Allthevalues were set to 0 if they were smaller than 0.5 and set to 1 otherwise in the predictions of the testing set before evaluatingthemetrics.
Volume:
Issue:
www.irjet.net

Each dilated inception block is followed by another dilatedinceptionblockuntilthebottlenecklayer.AMaxPooling layer is applied to the output from each block before passing it to the next one to reduce the dimensionsby half.Eachfour-pixel square ontheimage isreplacedbyMax-Poolingwithonepixelwhosevalueis equal tothe maximum ofthefourpixels.The numberof filtersineachblock isdoublethatoftheprevious block. The model is then enabled to learn more low-level features.Moreover,aresidualconnectionisaddedtothe model to pass the output from each of these blocks to their corresponding decoder blocks in the U-Net symmetryfortheretention ofspatial informationwhich the decoder blocks can further use to extract valuable features out of the image. The bottleneck layer which is capable of detecting rich but most spatially inaccurate features is where the encoding of the image to a lower scale cease. There are two bottleneck blocks, the first of whichpassesitsoutputwithoutchangingthedimension tothesecond.Theseblocksareexpectedtolearnawide varietyoflow-levelfeatures,hencetherearemorefilters intheseblocksascomparedtotheotherones.
The output from the decoder block contains useful information for the classification of pixels. The output’s shape must be expanded in order to align with the final output’s shape. The dimensions are enlarged using transposed convolutions that expand the output's size usingtrainablefilters.Incontrasttoamax-poolinglayer, this scales the output up to twice its original size and
consists of trainable parameters. Before passing this scaled-up information to the next block, the outputs fromthecorrespondingencoderblocksareconcatenated to the output using the residual (or skip) connections mentionedbefore.Thescaling-upisdoneuntilthesizeof the output acquired is the same as the input. A sigmoid activation layer, which is ideal for binary classification tasksisthenappliedtotheoutputfromthelastdecoding blocktofinallyclassifythepixel.
In the field of medical image analysis, merely an increase in accuracy cannot fully explain how well deep learning algorithms perform, where issues such as class disproportionindataandthecatastrophicrepercussions of skipped tests must be taken into account [15]. To evaluate the model, two metrics were used: DICE Coefficient and Aggregated Jaccard Index. DICE coefficient is defined as the ratio of twice the intersection between the ground truth and predicted mask to the sum of the ground truth and prediction masks(ref).InordertoevaluatetheDICEcoefficient,the pixelvaluesofthemaskswerescaleddownintherange between0and1.Theproductofthepredictedmaskand the ground truth would be equal to the area of intersection between the two. Summing the two image arrayswouldgivethetotalareaofbothimages.
whereGisthegroundtruthandPisthepredictedmask of the nuclei. The numerator term represents their
International Research Journal of Engineering and Technology (IRJET)

e-ISSN:2395-0056
Volume: 09 Issue: 10 | Oct 2022 www.irjet.net p-ISSN:2395-0072
intersection, whereas the denominator term is simply thesum oftheirareas.AggregatedJaccardIndex(AJI) is ametrictoevaluatesegmentationquality,definedasthe ratio of the intersection of the ground truth and predictedmasktothesumoftheirunion,falsepositives, andfalsenegatives.
whereSisthesetofallpixelsinthepredictedmaskand groundtruththatare mismatched thoseaccounts forall the false positives and false negatives. This score is naturally lower than the Jaccard Index or IoU as more values are being added to the denominator. This metric evaluates the performance of a model by penalizing for the mis predicted pixels, under-segmentation, and oversegmentation of nuclei. A DICE Coefficient-based loss functiondefinedasthenegativenaturallogarithmofthe DICECoefficientwasusedforthemodel.
Higher values of the DICE Coefficient imply a better matchofgroundtruthand predictedmask,sothelower values of the loss function indicate better segmentation quality.Therangeofthelossfunctionis[0,∞)where0is the absolute perfect case of ground truth and predicted mask matching with each other or them being exactly thesame.
The model’s architecture was constructed from scratch and was trained on the 480 original patches of the color normalized images and approximately 2500 augmented images using Adam Optimizer with a learning rate of0.001for20epochs with a batchsizeof 10 images and the model with the best validation DICE Coefficient was chosen and used for evaluation on the testingsetwhichwasused asvalidationdatainthefirst place. The entire training process took a little over an hour. The loss evaluated on the testing set was 0.2005. The overall segmentation quality was impressive considering the small training data used and the small training time. It was discovered that the model was relatively weaker at segmenting the nuclei belonging to the breast and colon images as is evident by the AJI scores on these organs. Table 1 shows the DICE Coefficient scores and AJI scores on different organs in thetestingdata.
Table -1:
Organ DICE AJI
Bladder 0.843 0.732
Brain 0.817 0.696 Breast 0.782 0.635 Colon 0.775 0.630 Kidney 0.824 0.700 Lung 0.827 0.702 Prostate 0.819 0.683
Results
Score
Loss 0.2005 AJI 0.6877 DICE 0.8183
The DICE Coefficient score and Aggregated Jaccard Index were evaluated to be 0.8183 and 0.6877 on the entiretestingset.Themodelwassuccessfulineffectively separatingthe boundariesofthe nuclei,and thedensely packedclustersofnucleiwereseparatedprettywell.Ina comparisonofthepredictedmasktothegroundtruth,a set of false-positive nuclei was discovered. The tissue images and their corresponding ground truth binary masks and predicted masks from the Dilated Inception U-Net model can be seen in Fig 6 This modified U-Net model used for nuclei segmentation was inspired by D Cahall’s[8]paperinwhichtheyusedthesamemodelfor brain tumor segmentation. There were specific changes madetothemodeltofitbettertothedatasetbeingdealt with. In contrast to the convention in U-Net models, anotherbottlenecklayerwasintroducedinsequencefor the extraction of a set of even more complex features from input images. The same loss function was adapted but for binary classification. It is worth noting that the Dilated Inception U-Net is a computationally efficient model with a much faster training time which is made possible due to the dilated convolutions. The blocks utilizing inception modules (non-dilated convolutions) would have to train an absurd number of parameters, whichwouldbemorethantwiceasmanyasthoseinthe dilated inception blocks if it weren't for the dilated convolutions. Besides the impressive scores on the testingdata,thereseemedtobeasubstantialnumberof false positives and false negatives in the predictions, which could be diminished using some post-processing techniques like image morphology or removal of extremely small predicted instances, leaving some groundforfutureresearch.
International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056

Volume: 09 Issue: 10 | Oct 2022 www.irjet.net p-ISSN:2395-0072
The most popular patch sizes have been 256x256 because they include more than enough contextual information from each nucleus' surroundings to extract meaningfulfeaturesforsegmentingthem.Asidefromthe massive amount of memory needed, feeding patches of thissizetothemodelassuresafasterpredictionandthe model hastolearna lotfewerparametersthanitwould if the original images of size 1000x1000 were used for training. Color normalization of the dataset before further processing has been a common method in past works of researchers. Impressive results were nonetheless obtained with color normalization and excessiveaugmentationapproacheswhichcompensated forstaininconsistenciesinthedataset.
The Dilated Inception U-Net model was used for nuclei segmentation in histopathology images. DICE coefficient and Aggregated Jaccard Index Score were used as evaluation metrics. Smaller image patches were extracted with minimum overlapping from each train and test image and multiple augmentation techniques were employed to improve the performance of the model. Despite the little amount of data provided, the model deliveredthe bestperformancesegmenting brain andbladdernuclei images. Furtherwork canbe doneto improve the performance of breast and colon nuclei segmentation by adopting different augmentation techniques. These sparse dilated filters were able to segmentthedenselypackednucleiinthetissuepictures, allowing the model to be generalizable to new unseen tissue images. The model showed promise in its generalizability by performing well on the unseen testingdatadespitebeingtrainedonasmalldataset.
[1] Cruz JA, Wishart DS. Applications of Machine Learning in Cancer Prediction and Prognosis. Cancer Informatics. January 2006. doi:10.1177/117693510600200030.
[2] Lei,T.,Wang,R.,Wan,Y.,Du,X.,Meng,H.andNandi, A.K., 2020. Medical Image Segmentation Using Deep Learning:ASurvey.
[3] N. Kumar et al., "A Multi-Organ Nucleus Segmentation Challenge," in IEEE Transactions on Medical Imaging, vol. 39, no. 5, pp. 1380-1391, May 2020,doi:10.1109/TMI.2019.2947628.
[4] N. Kumar, R. Verma, S. Sharma, S. Bhargava, A. Vahadane and A. Sethi, "A Dataset and a Technique for Generalized Nuclear Segmentation for Computational Pathology," in IEEE Transactions on Medical Imaging, vol. 36, no. 7, pp. 1550-1560, July 2017, doi: 10.1109/TMI.2017.2677499.
[5] Long, J., Shelhamer, E., and Darrell, T. (2015). “Fully convolutional networks for semantic segmentation,” in ProceedingsoftheIEEEConferenceonComputerVision and Pattern Recognition, (Boston, MA: IEEE), 3431–3440.doi:10.1109/ICCVW.2019.00113.
[6] V. Badrinarayanan, A. Kendall and R. Cipolla, "SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation," in IEEE Transactions on Pattern Analysis and Machine Intelligence,vol.39, no. 12, pp. 2481-2495,1 Dec. 2017, doi:10.1109/TPAMI.2016.2644615.
International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056

[7] Ronneberger, O., Fischer, P., Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N., Hornegger J., Wells W., FrangiA.(eds)MedicalImageComputingandComputerAssisted Intervention – MICCAI 2015. Lecture Notes in Computer Science, vol 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/9783-31924574_428.
[8] Z. Zeng, W. Xie, Y. Zhang and Y. Lu, "RIC-Unet: An Improved Neural Network Based on Unet for Nuclei Segmentation in Histology Images," in IEEE Access, vol. 7, pp. 21420-21428, 2019, doi: 10.1109/ACCESS.2019.2896920.
[9] Zhou, Yanning & Onder, Omer & Tsougenis, Efstratios & Chen, Hao & Kumar, Neeraj. (2020). MHFCN: Multi-Organ Nuclei Segmentation Algorithm. IEEE transactionsonmedicalimaging.
[10] M. Macenko, M. Niethammer, J. S. Marron, D. Borland, J. T. Woosley, X. Guan, C. Schmitt, and N. E. Thomas, “A method for normalizing histology slides for quantitative analysis,” in IEEE International Symposium on Biomedical Imaging: From Nano to Macro, 2009, pp. 1107–1110.
[11] Iqra Kiran, Basit Raza, Areesha Ijaz, Muazzam A. Khan, DenseRes-Unet: Segmentation of overlapped/clustered nuclei from multi organ histopathology images, Computers in Biology and Medicine, Volume 143, 2022, 105267, ISSN 0010-4825, https://doi.org/10.1016/j.compbiomed.2022.105267.
[12] Wang, Yunzhi & Kumar, Neeraj. (2020). Fully Convolutional Neural Network for Multi-Organ Nuclei Segmentation.IEEETransactionsonMedicalImaging.
[13] Kong,Y.,Genchev,G.Z.,Wang,X.,Zhao,H.andLu,H., 2020.NuclearSegmentationinHistopathologicalImages Using Two-Stage Stacked U-Nets With Attention Mechanism. Frontiers in Bioengineering and Biotechnology,p.1246.
[14] B.Menzeetal.,“TheMultimodalBrainTumorImage Segmentation Benchmark (BRATS),” IEEE Transactions on Medical Imaging, vol. 34, no. 10, pp. 1993–2024, Oct. 2014.(availableat:https://hal.inria.fr/hal-00935640).
[15] Cahall, D.E., Rasool, G., Bouaynaya, N.C. and Fathallah-Shaykh, H.M., 2021. Dilated Inception U-Net (DIU-Net)forBrainTumor Segmentation.arXivpreprint arXiv:2108.06772.
[16] “The cancer genome atlas (TCGA),” http://cancergenome.nih.gov/.
Volume: 09 Issue: 10 | Oct 2022 www.irjet.net p-ISSN:2395-0072 © 2022, IRJET | Impact Factor value: 7.529 | ISO 9001:2008 Certified Journal | Page 851
[17] Bradski, G. (2000). The OpenCV Library. Dr. Dobbs JournalofSoftwareTools.
[18] Abadi, Martín, et al. “TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems.” arXiv.Org, doi.org, 14 Mar. 2016, https://doi.org/10.48550/arXiv.1603.04467.
[19] C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S. Reed, D. Anguelov, D. Erhan, V. Vanhoucke, and A. Rabinovich. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition,pages1–9,2015.