EDITORIALS Fusion genes in acute myeloid leukemia: do acute myeloid leukemia diagnostics need to fuse with RNA-sequencing? Felicitas Thol Department of Hematology, Hemostasis, Oncology, and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany E-mail: FELICITAS THOL - thol.felicitas@mh-hannover.de doi:10.3324/haematol.2021.278983
I
n this issue of Haematologica, Kerbs and colleagues present a detailed analysis of fusion gene detection by RNAsequencing in acute myeloid leukemia (AML).1 Fusion genes, i.e., genes made by joining parts of two different genes, occur frequently in carcinogenesis. These fusion transcripts can occur within one chromosome as well as between two chromosomes (inter-chromosomal) and include translocations, deletions, insertions as well as inversions. Thousands of chromosomal aberrations and fusion genes have been identified as being recurrently present in cancer.2 Interestingly, in pediatric acute lymphocytic leukemia it was shown that many fusion genes are already present in utero and that a further transformation occurs in early childhood through secondary mutations.3 The discovery of BCR-ABL1 in patients with chronic myeloid leukemia as well as gene fusions involving neurotrophic tyrosine receptor kinase (NTRK)4 and neuregulin-1 (NRG1)5 in solid cancers are examples of how the identification of fusion genes have not only revolutionized the diagnosis but also the therapy of a disease. It is estimated that fusion genes occur in 30% of patients with AML6 and it has been recognized that certain recurrent chromosomal and genetic aberrations delineate distinct types of AML. Along this line, the current World Health Organization (WHO) classification of AML describes seven types of recurrent genetic abnormalities in AML as well as the provisional entity “AML with BCR-ABL1”.7 Furthermore, the WHO classification lists recurrent genetic aberrations that classify the disease as “AML with myelodysplasia-related changes”. This
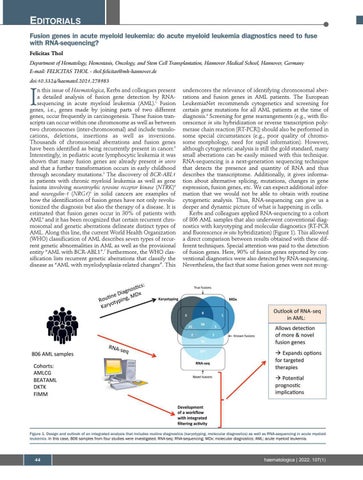
underscores the relevance of identifying chromosomal aberrations and fusion genes in AML patients. The European LeukemiaNet recommends cytogenetics and screening for certain gene mutations for all AML patients at the time of diagnosis.8 Screening for gene rearrangements (e.g., with fluorescence in situ hybridization or reverse transcription polymerase chain reaction [RT-PCR]) should also be performed in some special circumstances (e.g., poor quality of chromosome morphology, need for rapid information). However, although cytogenetic analysis is still the gold standard, many small aberrations can be easily missed with this technique. RNA-sequencing is a next-generation sequencing technique that detects the presence and quantity of RNA and thus describes the transcriptome. Additionally, it gives information about alternative splicing, mutations, changes in gene expression, fusion genes, etc. We can expect additional information that we would not be able to obtain with routine cytogenetic analysis. Thus, RNA-sequencing can give us a deeper and dynamic picture of what is happening in cells. Kerbs and colleagues applied RNA-sequencing to a cohort of 806 AML samples that also underwent conventional diagnostics with karyotyping and molecular diagnostics (RT-PCR and fluorescence in situ hybridization) (Figure 1). This allowed a direct comparison between results obtained with these different techniques. Special attention was paid to the detection of fusion genes. Here, 90% of fusion genes reported by conventional diagnostics were also detected by RNA-sequencing. Nevertheless, the fact that some fusion genes were not recog-
Figure 1. Design and outlook of an integrated analysis that includes routine diagnostics (karyotyping, molecular diagnostics) as well as RNA-sequencing in acute myeloid leukemia. In this case, 806 samples from four studies were investigated. RNA-seq: RNA-sequencing; MDx: molecular diagnostics; AML: acute myeloid leukemia.
44
haematologica | 2022; 107(1)